Modulators of Protein-Protein Interactions
Whilst the traditional targets for small molecule drug discovery have been enzymes, ion channels or receptors there has recently been an increase in interest in targeting protein-protein interactions (PPI). To date very few of the estimated > 650,000 PPI’s DOI are well characterised, they are thought to be involved in signal transduction, cell adhesion, cellular proliferation, growth, differentiation, viral self-assembly, programmed cell death, and cytoskeleton structure. They are also a critical component of a number of pathologies e.g. amyloidosis or prion diseases. It is also important to note that as well as inhibitors of protein-protein interactions small molecules have been identified that stabilise protein-protein interactions.
Part of the reason for the increased interest in PPI is due to the emergence of new biophysical technologies that allow study, many of which involve technologies used for fragment screening such as Capillary Electrophoresis , SPR, ITC together with advances in X-ray crystallography and NMR.
Forces involved in Protein-Protein Interactions
The contact surfaces between the proteins is relatively large (800 A2) but can vary (500-5000A2) Proc. Natl. Acad. Sci. USA 1996, 93, 13–20, steric factors, hydrophobic and electrostatic interactions and hydrogen bonds all contribute to the binding interaction however it has been shown that hydrophobic forces are significant.
Studies by Nussinov (Protein Engineering vol.10 no.9 pp.999–1012, 1997)
found that, on average, there are 10.7 hydrogen bonds and 2.0 salt bridges per interface. Charge complementarity is found for both charges and hydrogen bonding donors/acceptors. However, 17.4% of fully buried donors or acceptors in high- resolution structures do not form any hydrogen bonds, and some like charges are at a close distance. Polar atoms on the backbone have a strong tendency to form hydrogen bonds with backbone atoms across the interface, and some main chain–main chain hydrogen bonds can form β-sheets.
These results indicate that within the interface region, polar and charged residues constitute a larger percentage than would normally be found on the surface of proteins. Water molecules are often found buried within the interface and can form a network of hydrogen bonds that help stabilise the complex.
These results refer to the whole of the potential binding site, TIMBAL is a database containing small molecules that modulate protein-protein interactions, it also includes the PDB codes for a number of the records. If we use those 689 PDB records for which there is a ligand present we can calculate which residues of the protein are with 3A of the ligand using countpocketaa.svl script within MOE. The plot below created in Aabel scores each of the PDB structures for presence/absence of each of the amino acids.
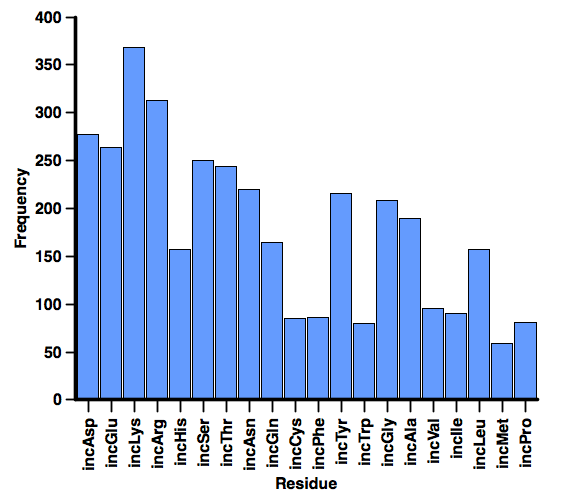
Interestingly the most common residues are the four ionisable residues, Asp, Glu, Lys and Arg, 404 of the proteins have an acidic residue within 3A of the ligand, and 588 have a basic amino acid. In addition 574 have an amino acid side-chain capable of making a hydrogen bond.
So whilst the interacting surfaces on the proteins may be large and hydrophobic surfaces may be significant it appears that almost all existing ligands bind close to charged residues.
Small molecule modulators
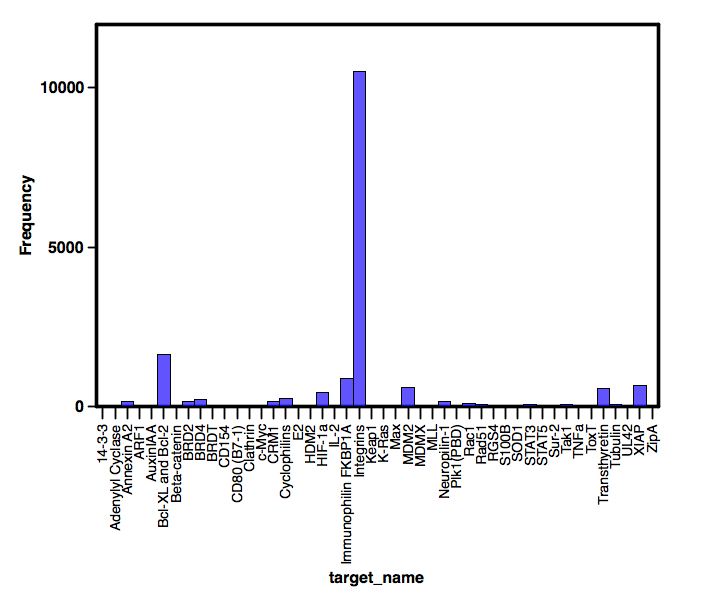
TIMBAL is a database containing small molecules that modulate protein-protein interactions, the structures are automatically extracted from ChEMBL using searches based on a list of known PPI targets. The database contains over 17,000 data points for 8,107 unique molecules that have been tested against 50 PPI targets, however not all targets are equally investigated with integrins in particular having many more molecules associated with it. In an ideal world all targets would be equally represented, in addition there is no information about the mode of interaction (reversible or irreversible), but these are early days and hand curation would be a huge task.
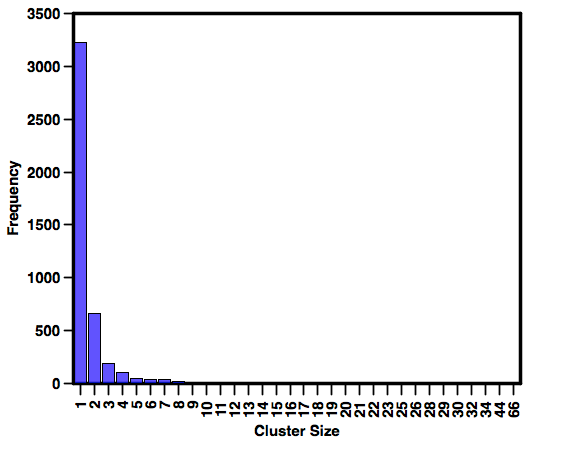
With the structures coming from a limited selection of targets there is also a concern about diversity. In order to get some idea of the diversity of the molecules I clustered them in MOE using MACCS fingerprints and a Tanimoto cutoff of 0.85, the chart below was then created in Aabel. Whilst there are a couple of large clusters (the 66 membered cluster targets Integrin), the vast majority of the compounds are singletons or only have one or two similar compounds. I addition it does seem, at least from this limited collection, that molecules for one target are not similar to molecules for another target.
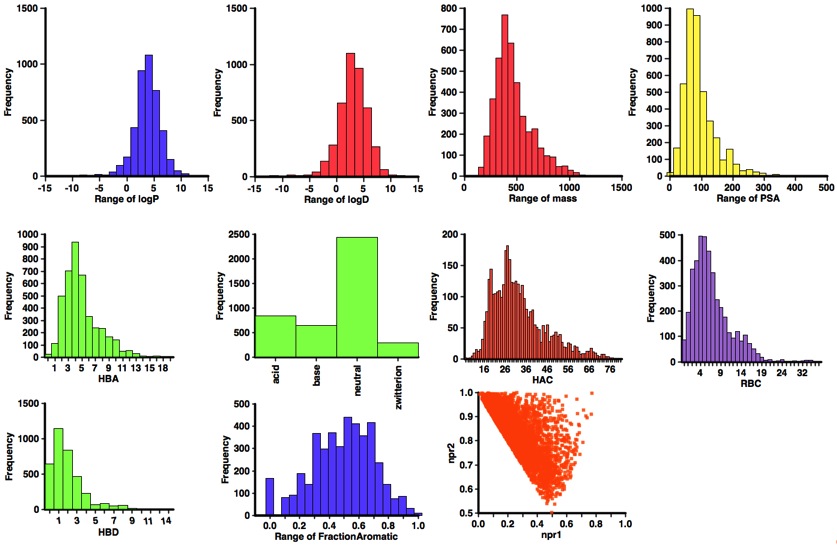
A profile of the physicochemical properties (HBD, HBA, PSA, HAC, LogP, LogD, MWt, RBC) of all molecules was generated using an Applescript that uses evaluate from ChemAxon to calculate the physicochemical properties and Aabel to construct the histograms. I also used it to determine pKa in order to identify acidic or basic groups and categorized the molecules accordingly, in addition I calculated the fraction of aromatic atoms (number of aromatic atoms/number of heavy atoms) since there has been concern about the number of aromatic compounds in sample collections. I’ve also included npri (Normalized ratio of principle moments of inertia) as described by Sauer WH, Schwarz MK (2003) Molecular shape diversity of combinatorial libraries: A prerequisite for broad bioactivity. J Chem Inf Comput Sci 43:987–10030. DOI this was calculated using MOE.
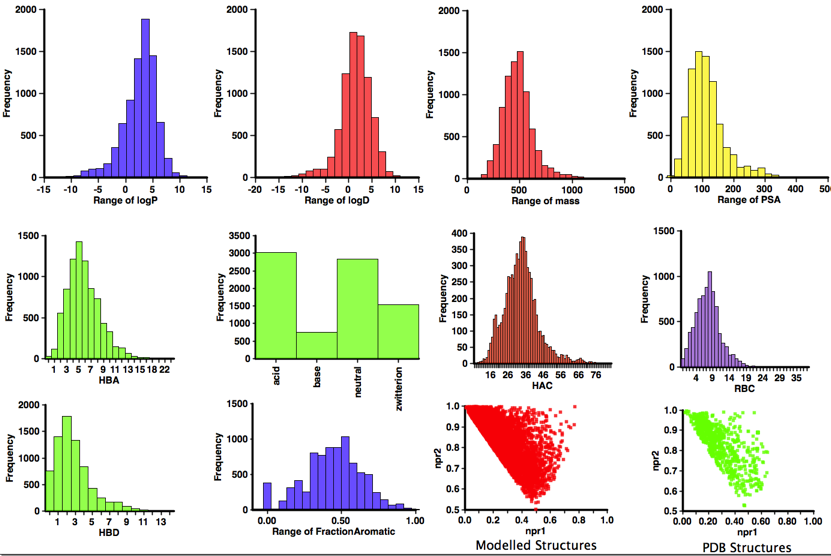
As one might have expected the structures are rather large, with many having a molecular weight >500. However the lipophilicity and especially the LogD is in general in an acceptable range, this is because many of the ligands contain an ionisable group. Indeed only 30% of the ligands are predicted to be neutral at physiological pH. Whilst there may be concerns that many of these structures are not rule-of-five compliant it is important to note that a number look to be derived from natural products, and Lipinski (Advanced Drug Delivery Reviews 46 (2001) 3 – 26) excluded natural products from his analysis. All structures passed the PAINS filter DOI.
The 3D shape measures (npr1 and npr2) are interesting, with a concentration along the rod-disc axis but with a few structures populated the 3D space, however the compounds contain up to 20 rotatable bonds and so taking a single minimised structure may not give a good impression of the 3D shape. However for around 1000 of the structures the ligand bound conformation is available from the PDB, and so I’ve calculated npr1 and npr2 for these conformations and plotted the results alongside those for the modelled structures. Again we see that the more 3D space is sparsely populated.
What is particularly striking is that nearly 60% of the ligands contain an acidic group, closer inspection reveals that this is due to the molecules acting on Integrins, if they are removed (see plot below) from the analysis then the number of structures containing an acid is reduced and the number of neutral molecules increases to just over 50%.
Conclusions
Admittedly this is a relatively small dataset but it does look like that each protein-protein interaction will require a different chemotype. The molecules are rather larger than would be expected from a traditional drug discovery program, and whilst the PPI site is large and hydrophobic forces are significant, it would be better to target ionisable residues and thus include an increased proportion of acids and bases in any screening collection.
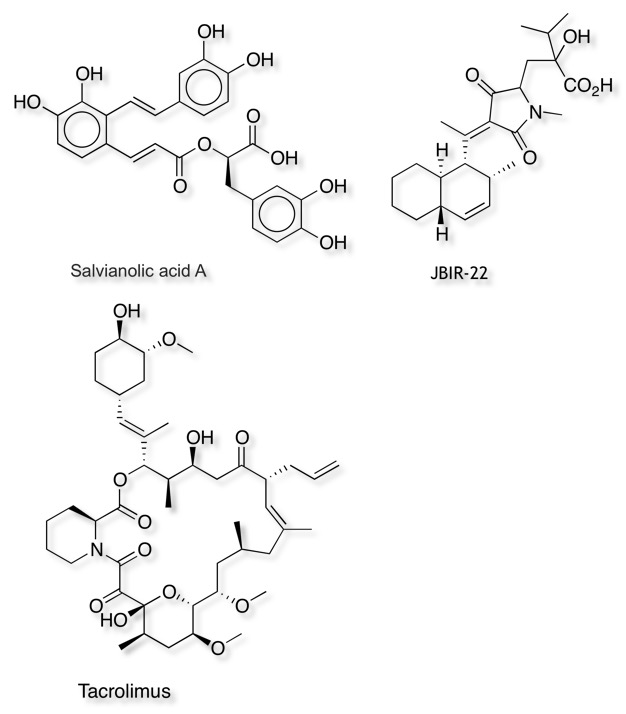
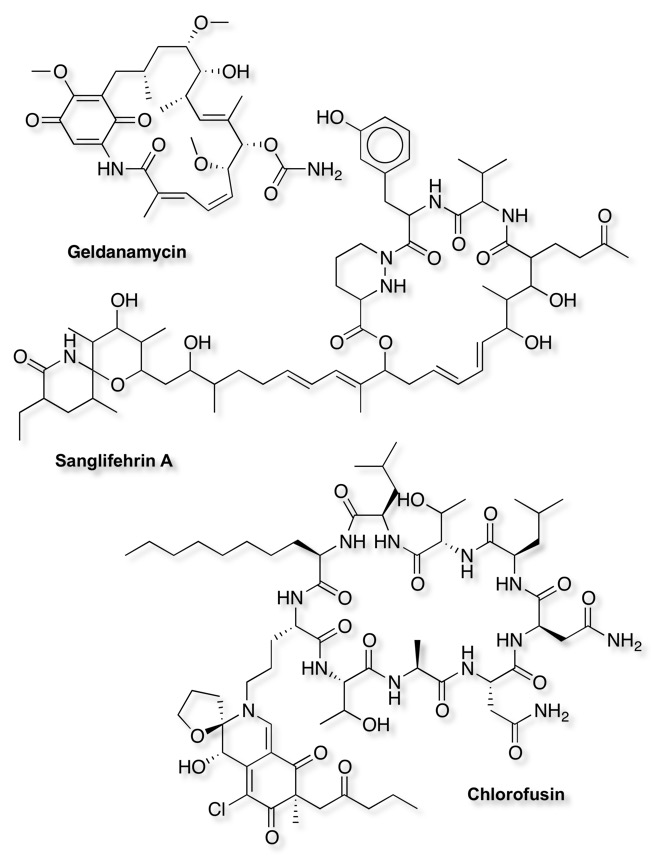
As I mentioned a number of the known ligands look like they are natural products or are derived from natural products, and indeed there are number of important examples of natural products involved in PPI, Sanglifehrins for Cyclophilins, geldanomycin for HSP90, chlorofusin MDM1-p53. An attractive approach might be to search a natural product library, and as I mentioned above Natural Products were not part of the Lipinski evaluation, and the internal hydrogen bonds etc. can lead to surprising pharmacokinetics. Some examples are shown below.
. 
Updated 30 August 2013