Examples of Fingerprint and Descriptors
Fingerprints or descriptors are an abstract representation of certain structural features of a molecule. These descriptors may represent a structural key within a molecule. This might be as simple as a count of a particular atom type, S, N etc, or halogen, or sp3. It might be the presence of a particular ring system e.g. Phenyl, Pyridyl, Naphthyl, or a functional group e.g. Amide, Ester, Amine. It might be a calculated property Hydrogen Bond donor, Polar Surface area, LogP. Fingerprints are more abstract than a structural key but have the advantage of being more general since they do not represent pre-defined patterns.
Unlike a structural key with its pre-defined patterns, the patterns for a molecule's fingerprint are generated from the molecule itself. The fingerprinting algorithm examines the molecule and generates the fingerprint based on a set of rules.
Fingerprints
Path-based fingerprints FP2, a path-based fingerprint which indexes small molecule fragments based on linear segments of up to 7 atoms. A molecule structure is analysed to identify linear fragments of length from 1-7 atoms. Single atom fragments of C, N, and O are ignored. A fragment is terminated when the atoms form a ring. For each of these fragments the atoms, bonding and whether they constitute a complete ring is recorded and saved in a set so that there is only one of each fragment type. Chemically identical versions, (i.e. ones with the atoms listed in reverse order and rings listed starting at different atoms) are identified and only a single canonical fragment is retained. Each remaining fragment is assigned a hash number from 0 to 1020 which is used to set a bit in a 1024 bit vector.
Atom Pairs, shortest path between all pairs of atoms ref
Topological Torsion, the TT consists of four consecutive bonded non-hydrogen atoms ref
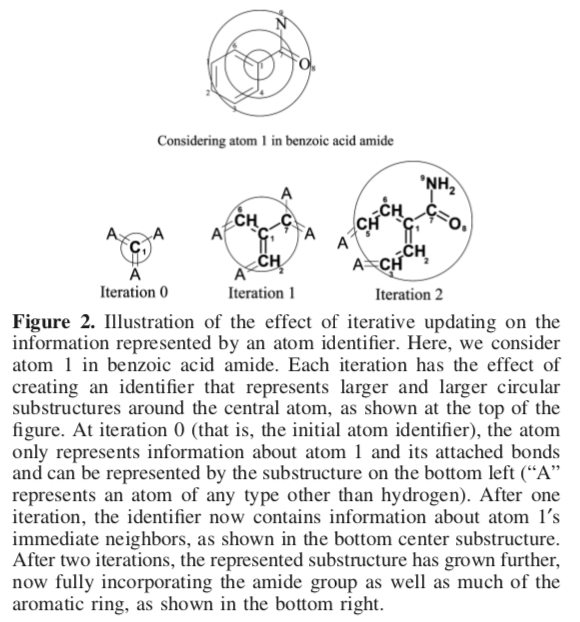
Extended Connectivity Fingerprints (ECFPs, or "Circular Fingerprints") DOI offer a number of advantages over other schemes.
ECFPs were developed specifically for structure−activity modeling. ECFPs are circular fingerprints with a number of useful qualities: they can be very rapidly calculated; they are not predefined and can represent an essentially infinite number of different molecular features (including stereochemical information); their features represent the presence of particular substructures, allowing easier interpretation of analysis results; and the ECFP algorithm can be tailored to generate different types of circular fingerprints, optimized for different uses.
There is an excellent description of ECFP in a recent blog post.
There are also fingerprints using SMARTS patterns.These generate a topological fingerprint for a molecule using a series of pre-defined structural patterns FP4,
Some examples of descriptors that can be calculated using MOE
a_aro Number of aromatic atoms.
a_count Number of atoms (including implicit hydrogens).
This is calculated as the sum of (1 + hi) over all non-trivial atoms i.
a_heavy Number of heavy atoms #{Zi | Zi > 1}.
a_ICM Atom information content (mean).
This is the entropy of the element distribution in the molecule (including implicit hydrogens but not lone pair pseudo-atoms).
Let ni be the number of occurrences of atomic number i in the molecule.
Let pi = ni / n where n is the sum of the ni.
The value of a_ICM is the negative of the sum over all i of pi log pi.
a_IC Atom information content (total).
This is calculated to be a_ICM times n.
a_nH Number of hydrogen atoms (including implicit hydrogens).
This is calculated as the sum of hi over all non-trivial atoms i plus the number of non-trivial hydrogen atoms.
a_nB Number of boron atoms: #{Zi | Zi = 5}.
a_nC Number of carbon atoms: #{Zi | Zi = 6}.
a_nN Number of nitrogen atoms: #{Zi | Zi = 7}.
a_nO Number of oxygen atoms: #{Zi | Zi = 8}.
a_nF Number of fluorine atoms: #{Zi | Zi = 9}.
a_nP Number of phosphorus atoms: #{Zi | Zi = 15}.
a_nS Number of sulfur atoms: #{Zi | Zi = 16}.
a_nCl Number of chlorine atoms: #{Zi | Zi = 17}.
a_nBr Number of bromine atoms: #{Zi | Zi = 35}.
a_nI Number of iodine atoms: #{Zi | Zi = 53}.
b_1rotN Number of rotatable single bonds.
Conjugated single bonds are not included (e.g. ester and peptide bonds).
b_ar Number of aromatic bonds.
b_count Number of bonds (including implicit hydrogens).
This is calculated as the sum of (di/2 + hi) over all non-trivial atoms i.
b_double Number of double bonds.
Aromatic bonds are not considered to be double bonds.
b_heavy Number of bonds between heavy atoms.
b_rotN Number of rotatable bonds.
A bond is rotatable if it has order 1, is not in a ring, and has at least two heavy neighbors.
b_single Number of single bonds (including implicit hydrogens).
Aromatic bonds are not considered to be single bonds.
b_triple Number of triple bonds.
Aromatic bonds are not considered to be triple bonds.
chiral The number of chiral centers.
lip_acc The number of O and N atoms.
lip_don The number of OH and NH atoms.
lip_druglike One if and only if lip_violation < 2 otherwise zero.
lip_violation The number of violations of Lipinski's Rule of Five
nmol The number of molecules (connected components).
opr_brigid The number of rigid bonds from [Oprea 2000].
opr_leadlike One if and only if opr_violation < 2 otherwise zero.
opr_nring The number of ring bonds from [Oprea 2000].
opr_nrot The number of rotatable bonds from [Oprea 2000].
opr_violation The number of violations of Oprea's lead-like test [Oprea 2000].
rings The number of rings.
Pharmacophore type counts
a_acc Number of hydrogen bond acceptor atoms (not counting acidic atoms but counting atoms that are both hydrogen bond donors and acceptors such as -OH).
a_acid Number of acidic atoms.
a_base Number of basic atoms.
a_don Number of hydrogen bond donor atoms (not counting basic atoms but counting atoms that are both hydrogen bond donors and acceptors such as -OH).
a_hyd Number of hydrophobic atoms.
vsa_acc Approximation to the sum of VDW surface areas (√Ö2) of pure hydrogen bond acceptors (not counting acidic atoms and atoms that are both hydrogen bond donors and acceptors such as -OH).
vsa_acid Approximation to the sum of VDW surface areas of acidic atoms (√Ö2).
vsa_base Approximation to the sum of VDW surface areas of basic atoms (√Ö2).
vsa_don Approximation to the sum of VDW surface areas of pure hydrogen bond donors (not counting basic atoms and atoms that are both hydrogen bond donors and acceptors such as -OH) (√Ö2).
vsa_hyd Approximation to the sum of VDW surface areas of hydrophobic atoms (√Ö2).
vsa_other Approximation to the sum of VDW surface areas (√Ö2) of atoms typed as "other".
vsa_pol Approximation to the sum of VDW surface areas (√Ö2) of polar atoms (atoms that are both hydrogen bond donors and acceptors), such as -OH
Charge based
ASA+ Water accessible surface area of all atoms with positive partial charge (strictly greater than 0).
ASA- Water accessible surface area of all atoms with negative partial charge (strictly less than 0).
ASA_H Water accessible surface area of all hydrophobic (|qi|<0.2) atoms.
ASA_P Water accessible surface area of all polar (|qi|>=0.2) atoms.
DASA Absolute value of the difference between ASA+ and ASA-.
CASA+ Positive charge weighted surface area, ASA+ times max { qi > 0 } [Stanton 1990].
CASA- Negative charge weighted surface area, ASA- times max { qi < 0 } [Stanton 1990].
DCASA Absolute value of the difference between CASA+ and CASA- [Stanton 1990].
dipole Dipole moment calculated from the partial charges of the molecule.
Examples available using ChemAxon cxcalc
atomcount, composition, dotdisconnectedformula, dotdisconnectedisotopeformula, elemanal, elementalanalysistable, exactmass, formula, icomposition, iformula, isotopecomposition, isotopeformula, mass
Charge atomicpolarizability, atompol, averagemolecularpolarizability, averagepol, avgpol, axxpol, ayypol, azzpol, charge, formalcharge, ioncharge, molecularpolarizability, molpol, oen, orbitalelectronegativity, pol, polarizability, tholepolarizability, tpol, tpolarizability
Conformation conformers, hasvalidconformer, leconformer, lowestenergyconformer, moldyn, moleculardynamics
Geometry aliphaticatom, aliphaticatomcount, aliphaticbondcount, aliphaticringcount, aliphaticringcountofsize, angle, aromaticatom, aromaticatomcount, aromaticbondcount, aromaticringcount, aromaticringcountofsize, asa, asymmetricatom, asymmetricatomcount, asymmetricatoms, balabanindex, bondcount, bondtype, carboaliphaticringcount, carboaromaticringcount, carboringcount, chainatom, chainatomcount, chainbond, chainbondcount, chiralcenter, chiralcentercount, chiralcenters, connected, connectedgraph, cyclomaticnumber, dihedral, distance, distancedegree, dreidingenergy, eccentricity, fragmentcount, fusedaliphaticringcount, fusedaromaticringcount, fusedringcount, hararyindex, heteroaliphaticringcount, heteroaromaticringcount, heteroringcount, hindrance, hyperwienerindex, largestatomringsize, largestringsize, largestringsystemsize, maximalprojectionarea, maximalprojectionradius, maximalprojectionsize, minimalprojectionarea, minimalprojectionradius, minimalprojectionsize, molecularsurfacearea, msa, plattindex, polarsurfacearea, psa, randicindex, ringatom, ringatomcount, ringbond, ringbondcount, ringcount, ringcountofatom, ringcountofsize, ringsystemcount, ringsystemcountofsize, rotatablebond, rotatablebondcount, shortestpath, smallestatomringsize, smallestringsize, smallestringsystemsize, stereodoublebondcount, stericeffectindex, sterichindrance, szegedindex, topanal, topologyanalysistable, vdwsa, volume, wateraccessiblesurfacearea, wienerindex, wienerpolarity
Isomers canonicaltautomer, dominanttautomerdistribution, doublebondstereoisomercount, doublebondstereoisomers, generictautomer, majortautomer, moststabletautomer, stereoisomercount, stereoisomers, tautomercount, tautomers, tetrahedralstereoisomercount, tetrahedralstereoisomers
Markush Enumerations enumerationcount, enumerations, markushenumerationcount, markushenumerations, randommarkushenumerations
Partitioning logd, logp
Protonation averagemicrospeciescharge, chargedistribution, isoelectricpoint, majormicrospecies, majorms, microspeciesdistribution, msdistr, pi, pka
Other acc, acceptor, acceptorcount, acceptormultiplicity, acceptorsitecount, acceptortable, accsitecount, aromaticelectrophilicityorder, aromaticnucleophilicityorder, canonicalresonant, chargedensity, don, donor, donorcount, donormultiplicity, donorsitecount, donortable, donsitecount, electrondensity, electrophilicityorder, electrophiliclocalizationenergy, frameworks, hbda, hbonddonoracceptor, hmochargedensity, hmoelectrondensity, hmoelectrophilicityorder, hmoelectrophiliclocalizationenergy, hmohuckel, hmohuckeleigenvalue, hmohuckeleigenvector, hmohuckelorbitals, hmohuckeltable, hmolocalizationenergy, hmonucleophilicityorder, hmonucleophiliclocalizationenergy, hmopienergy, huckel, huckeleigenvalue, huckeleigenvector, huckelorbitals, huckeltable, localizationenergy, msacc, msdon, nucleophilicityorder, nucleophiliclocalizationenergy, pichargedensity, pienergy, refractivity, resonantcount, resonants, totalchargedensity
Examples from OpenBabel
name [Name]
formula [Formula]
molweight [Molecular Weight]
exactmass [Isotopic Mass]
canonicalSMILES [String]
numatoms [Number]
numbonds [Number]
numresidues [Number]
sequence [Residue Sequence]
num_rings [Number of Rings (by SSSR)]
logP [Number (octanol-water partition)]
PSA [Number (topological polar surface area)]
MR [Number (molar refractivity)
Examples from MayaChemTools
MolecularWeight, ExactMass, HeavyAtoms, Rings, AromaticRings, van der Waals MolecularVolume [ Ref 93 ], RotatableBonds, HydrogenBondDonors, HydrogenBondAcceptors, LogP and Molar Refractivity (SLogP and SMR), Topological Polar Surface Area (TPSA), Fraction of SP3 carbons (Fsp3Carbons) and SP3 carbons (Sp3Carbons), MolecularComplexity
Examples from Filter-it
element rules
topological property rules
ATOMS
CARBONS
HETEROATOMS
HETEROCARBONRATIO
HALIDES
HALIDEFRACTION
BONDS
ROTATABLEBONDS
RIGIDBONDS
FLEXIBILITY
CHIRALCENTERS
HBONDACCEPTORS
HBONDDONORS
LIPINSKIACCEPTORS
LIPINSKIDONORS
FORMALCHARGES
TOTALFORMALCHARGE
RINGS
ATOMSINSMALLESTRING
ATOMSINLARGESTRING
RINGFRACTION
AROMATICRINGS
ATOMSINSMALLESTAROMATICRING
ATOMSINLARGESTAROMATICRING
AROMATICRINGFRACTION
AROMATICOVERTOTALRINGFRACTION
NONAROMATICRINGS
ATOMSINSMALLESTNONAROMATICRING
ATOMSINLARGESTNONAROMATICRING
NONAROMATICRINGFRACTION
RINGSYSTEMS
ATOMSINSMALLESTRINGSYSTEM
ATOMSINLARGESTRINGSYSTEM
RINGSYSTEMFRACTION
RINGSINSMALLESTRINGSYSTEM
RINGSINLARGESTRINGSYSTEM
SIDECHAINS
ATOMSINSMALLESTSIDECHAIN
ATOMSINLARGESTSIDECHAIN
SIDECHAINFRACTION
CORES
ATOMSINCORE
COREFRACTION
BRIDGES
ATOMSINSMALLESTBRIDGE
ATOMSINLARGESTBRIDGE
BRIDGEFRACTION
physical property rules
MOLWT
LOGP
LOGS
TPSA
ANDREWSENERGY
LIGANDEFFICIENCY
fragment rules
Andrews Energy
Ligand Efficiency
ADMETSCORE
LIPINSKI_VIOLATIONS
ABSORPTION
PaDEL
PaDEL‐Descriptor is a software for calculating molecular descriptors and fingerprints DOI. The software currently calculates 797 descriptors (663 1D, 2D descriptors, and 134 3D descriptors) and 10 types of fingerprints.
| Descriptor class | Descriptor typea | Number of descriptors | Calculation speed (mol/s)b |
|---|---|---|---|
| ALOGP | 3 | 1084 | |
| APol | 1 | 24,738 | |
| Aromatic atoms count | 1 | 16,878 | |
| Aromatic bonds count | 1 | 16,336 | |
| Atom count | 13 | 2127 | |
| Autocorrelation (charge) | 5 | 6215 | |
| Autocorrelation (mass) | 5 | 777 | |
| Autocorrelation (polarizability) | 5 | 741 | |
| BCUT | 6 | 653 | |
| Bond count | 5 | 6014 | |
| BPol | 1 | 23,060 | |
| Carbon types | 9 | 20,327 | |
| Chi chain | 10 | 310 | |
| Chi cluster | 8 | 439 | |
| Chi path | 16 | 310 | |
| Chi path cluster | 6 | 347 | |
| Eccentric connectivity index | 1 | 11,611 | |
| Atom type electrotopological state | 482 | 289 | |
| Fragment complexity | 1 | 27,400 | |
| Hbond acceptor count | 1 | 16,126 | |
| Hbond donor count | 1 | 16,384 | |
| Kappa shape indices | 3 | 2413 | |
| Largest chain | 1 | 10,088 | |
| Largest Pi system | 1 | 13,804 | |
| Longest aliphatic chain | 1 | 10,233 | |
| Mannhold LogP | 1 | 16,328 | |
| McGowan volume | 1 | 546 | |
| Molecular distance edge | 19 | 2194 | |
| Molecular linear free energy relation | 6 | 300 | |
| Petitjean number | 1 | 10,131 | |
| Ring count | 34 | 1757 | |
| Rotatable bonds count | 1 | 11,471 | |
| Rule of five | 1 | 807 | |
| Topological polar surface area | 1 | 3535 | |
| Vertex adjacency information (magnitude) | 1 | 26,160 | |
| Weight | 1 | 24,532 | |
| Weighted path | 5 | 513 | |
| Wiener numbers | 2 | 10,755 | |
| XlogP | 1 | 910 | |
| Zagreb index | 1 | 23,593 | |
| Charged partial surface area | 29 | 309 | |
| Gravitational index | 9 | 10,012 | |
| Length over breadth | 2 | 8459 | |
| Moment of inertia | 7 | 9894 | |
| Petitjean shape index | 2 | 8213 | |
| WHIM (atomic masses) | 17 | 8229 | |
| WHIM (atomic polarizabilities) | 17 | 8150 | |
| WHIM (Mulliken atomic electronegativities) | 17 | 8290 | |
| WHIM (unit weights) | 17 | 8396 | |
| WHIM (van der Waals volumes) | 17 | 8072 | |
| CDK fingerprint | 1024 | 203 | |
| CDK extended fingerprint | 1024 | 189 | |
| CDK graph only fingerprint | 1024 | 223 | |
| Estate fingerprint | 79 | 276 | |
| MACCS fingerprint | 166 | 170 | |
| Pubchem fingerprint | 881 | 56 | |
| Substructure fingerprint | 307 | 116 | |
| Substructure fingerprint count | 307 | 113 | |
| Klekota‐Roth fingerprint | 4860 | 12 | |
| Klekota‐Roth fingerprint count | 4860 | 12 |
Updated 9 February 2019