Seasons Greetings
I hope you all have a great seasonal holiday, it has been a tough few years for folks so I think everyone needs a break. As Bad Company sang in Wishing Well.
But I know what you're wishing for, Love in a peaceful world
As in previous years rather post cards to everyone, instead any monies saved have been donated to the Multiple Sclerosis Society.

SureChEMBL Updated
SureChEMBL is a database of automatically abstracted patents, it uses three different methods to get structures, name to structure, image to structure and for some patents mol files if available. If you use it regularly you will be aware that it has become somewhat unreliable and the performance is not ideal.
This has just changed with an updated version of SureChEMBL.
Almost 10 years ago, EMBL-EBI acquired the SureChem system of chemically annotated patents and made this freely accessible in the public domain as SureChEMBL. Since then, our team has continued to maintain and deliver SureChEMBL. However, this has become increasingly challenging due to the complexities of the underlying codebase. We were awarded a Wellcome Trust grant in 2021 to completely overhaul SureChEMBL, with a new UI, backend infrastructure, and new features. We are now able to make available the first outputs from this project, which addresses the first two of these deliverables, with more to come in the future!
The new interface is here https://www.surechembl.org

If you have any issues you can submit them on GitHub https://github.com/chembl/surechembl-issues/issues.
One particularly useful new feature is the new public api. https://www.surechembl.org/api/swagger-ui.html. I'll certainly be exploring this in the future.
Molecular Interactions Updated
I've updated the Molecular Interactions page on the Drug Discovery Resources site.
https://www.cambridgemedchemconsulting.com/resources/molecular_interactions.html
Separation of PK and PD
Just catching up with my reading, I've always been a fan of compounds with slow off-rates and the impact on duration of action.
The situation was elegantly summarised in a publication from earlier in the year from Copeland et al. DOI
A dominant assumption in pharmacology throughout the 20th century has been that in vivo target occupancy-and attendant pharmacodynamics-depends on the systemic concentration of drug relative to the equilibrium dissociation constant for the drug-target complex. In turn, the duration of pharmacodynamics is temporally linked to the systemic pharmacokinetics of the drug. Yet, there are many examples of drugs for which pharmacodynamic effect endures long after the systemic concentration of a drug has waned to (equilibrium) insignificant levels. To reconcile such data, the drug-target residence time model was formulated, positing that it is the lifetime (or residence time) of the binary drug-target complex, and not its equilibrium affinity per se, that determines the extent and duration of drug pharmacodynamics.
I've added it to the page on separation of PK and PD in the Drug Discovery Resources
AlphaFold Protein Structure Database in 2024
A recent publication describes the continued evolution of the AlphaFold Protein Structure Database created by EMBL-EBI and DeepMind. From an initial 300K structures it now contains 214 million predicted protein structures.
You can read the paper here DOI.
The AlphaFold Database Protein Structure Database (AlphaFold DB, https://alphafold.ebi.ac.uk) has significantly impacted structural biology by amassing over 214 million predicted protein structures, expanding from the initial 300k structures released in 2021. Enabled by the groundbreaking AlphaFold2 artificial intelligence (AI) system, the predictions archived in AlphaFold DB have been integrated into primary data resources such as PDB, UniProt, Ensembl, InterPro and MobiDB. Our manuscript details subsequent enhancements in data archiving, covering successive releases encompassing model organisms, global health proteomes, Swiss-Prot integration, and a host of curated protein datasets. We detail the data access mechanisms of AlphaFold DB, from direct file access via FTP to advanced queries using Google Cloud Public Datasets and the programmatic access endpoints of the database. We also discuss the improvements and services added since its initial release, including enhancements to the Predicted Aligned Error viewer, customisation options for the 3D viewer, and improvements in the search engine of AlphaFold DB.
BMCS Hall of Fame
The BMCS have announced that nomination is open for the 2024 Hall of Fame award.
Details are here https://www.rscbmcs.org/awards/halloffame/.
Previous winners include
Dr Karin Briner, Professor Dame Carol Robinson, Dr David Rees, Sir Simon Campbell, Professor C Robin Ganellin,
EU restrictions on Fluorochemicals
In drug discovery the introduction of fluorine into potential drug candidates is an essential part of the medicinal chemists toolbox. Blocking a metabolic hotspot by replacing a Hydrogen by Fluorine or deactivating and aromatic ring by adding a trifluoromethyl are well established strategies for reducing metabolism, increasing half-life and reducing drug load. There are more details on the metabolism page and influence [pi-stacking interactionshttps://www.cambridgemedchemconsulting.com/resources/molecular_interactions.html).
I have a database of all drugs that have been reported to be in phase 3 trials or later (not all will have made it to market) and it is interesting to see how many contain a fluorine atom of some kind. As expected Aryl F and CF3 are the most common

A few examples are shown below.

There are also other classes of molecules like perflexane which is used as an imaging contrast agent in echocardiogram. Halothane used clinically as an inhalational anesthetic (on the WHO Model List of Essential Medicines) and other inhaled anesthetics.

So at first sight the EFMC statement was of some concern.
EFMC STATEMENT ON EU PROPOSAL TO BAN PFAS The EU, through the European Chemicals Agency (ECHA), has launched a proposal that aims to extensively restrict the manufacture, supply, and use of all per- and polyfluoroalkyl substances (Ref 1). PFAS are a large class of synthetic chemicals that are used across a broad range of activities in different scientific areas, not restricted to chemistry. However, several PFAS are environmental pollutants and some of them have detrimental effects on human health. Their extensive use throughout the society, combined with the low reactivity displayed by many fluoroalkyl chemicals, magnifies the potential for accumulation in the environment and contamination of food and water supplies. According to this proposal, which is in public consultation until September 25th (ref 2), PFAS encompasses "any substance that contains at least one fully fluorinated methyl (CF3) or methylene (CF2) without any H, Cl, Br, or I attached to it"
However, looking at the generic scope in more detail it appears that the scope might not be as all encompassing. Looking at the description here https://echa.europa.eu/registry-of-restriction-intentions/-/dislist/details/0b0236e18663449b. The generic scope is described as shown below and I've highlighted a critical phrase.
Per- and polyfluoroalkyl substances (PFASs) defined as: Any substance that contains at least one fully fluorinated methyl (CF3-) or methylene (-CF2-) carbon atom (without any H/Cl/Br/I attached to it).
A substance that only contains the following structural elements is excluded from the scope of the proposed restriction:
CF3-X or X-CF2-X’,
where X = -OR or -NRR’ and X’ = methyl (-CH3), methylene (-CH2-), an aromatic group, a carbonyl group (-C(O)-), -OR’’, -SR’’ or –NR’’R’’’,
and where R/R’/R’’/R’’’ is a hydrogen (-H), methyl (-CH3), methylene (-CH2-), an aromatic group or a carbonyl group (-C(O)-).
My interpretation (caveat I'm not a lawyer) is that the vast majority of drugs, reagents and solvents like trifluoro acetic acid would be excluded. However, inhaled anesthetics and the contrast imaging agents would be included. So the pharma industry needs to be included in the dialog but drugs themselves might have limited impact.
I've not covered polymers, and PTFE is is used in much laboratory equipment from stirrer bars, stopcocks, O-rings, seals etc. and who hasn't used PTFE tape on ground glass joints. There may be alternatives that I'm not aware of, but any substance that has similar properties of inertness, durability etc. may cause the same concerns as PFAs.
The European Chemicals Agency invites interested parties to send in scientific and technical information on the manufacture, placing on the market and use of per- and polyfluoroalkyl substances (PFAS) by 25 September 2023 https://echa.europa.eu/-/echa-seeks-input-on-proposed-pfas-restriction.
Diamond Light Source facility will be upgraded through a £500 million investment.
Diamond Light Source science facility in Oxfordshire will be upgraded through a £500 million investment.

Established in 2001 and opened in 2007, Diamond is a joint-venture between UKRI-STFC (86%) and the Wellcome Trust (14%).
The Diamond-II upgrade will take several years of planning and implementation. This will include a ‘dark period’ of 18 months with no synchrotron light for the user community, followed by a period to fully launch the new facility with three new flagship beamlines and major upgrades to many other beamlines.
£13 million for 22 AI for health research projects
UKRI have announced £13 invested in medical research projects. The projects aim to transform health using artificial intelligence (AI) to assist and refine diagnostics and procedures
https://www.ukri.org/news/13-million-for-22-ai-for-health-research-projects/
Includes image analysis in oncology, keyhole surgery, NLP analysis of clinical data and treatments for chronic pain.
UKQSAR meeting University of Liverpool on September 14th 2023.
The next UKQSAR meeting will be held at the University of Liverpool on September 14th 2023.
The meeting will take place at building 502, University of Liverpool (see D6 in campus map).
More information will be available on the website https://ukqsar.org/index.php/2023/07/19/uk-qsar-autumn-2023-meeting/
The schedule will be:
9.00-10.00 Registration (coffee/tea/refreshments) 10.00-10.15 Welcome – Nathan Brown (UK QSAR Chair)
10.15-11.45 Session 1 – Chair Neil Berry (University of Liverpool) 10.15-10.45 Talk 1– Alessandro Troisi (University of Liverpool) 10.45-11.15 Talk 2 – Abbie Trewin (University of Lancaster) 11.15-11.45 Talk 3 – Lauren Reid (Medchemica Ltd.) 11.45-12.45 Lunch and poster session
12.45-13.45 Session 2 – Chair Andrew Leach (University of Manchester) 12.45-13.15 Talk 4 – Steve Enoch (Liverpool John Moores University) 13.15-13.45 Talk 5 – Rachel Pirie (University of Newcastle) 13.45-14.15 Break (coffee/tea/refreshments) and poster session
14.15-15.45 Session 3 – Chair Steve Maginn (CCG) 14.15-15.15 Talk 6 – Elena De Orbe (AstraZeneca Ltd.) 15.15-15.45 Talk 7 – Adam Nelson (University of Leeds) 15.45-16.00 Conclusion and poster prizes - Nathan Brown (UK QSAR Chair)
Please register using the online form.
We are delighted that the RSC CICAG group (Chemical Information and Computer Applications Group) have been able to offer two travel bursaries for students to attend the meeting in return for providing a short write-up for the group. If you would like to apply for the bursary please use the form
Major user experience update in AlphaFold Database
Just saw this.
The AlphaFold Protein Structure Database, a result of a collaborative effort between Google DeepMind and EMBL’s European Bioinformatics Institute (EMBL-EBI), has released an exciting update to its web pages, providing users with an enhanced experience. This update marks a significant step in facilitating the use of AlphaFold structure data.
One of the most interesting updates are the improvements to the 3D viewer Mol*.
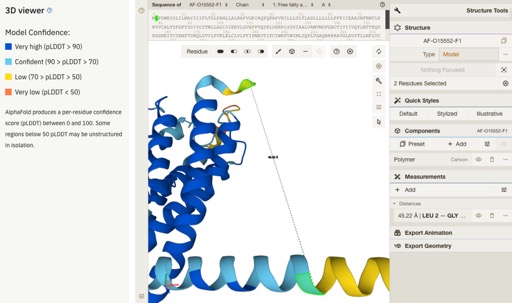
Full details are here https://www.ebi.ac.uk/about/news/updates-from-data-resources/alphafold-database-ux-update/.
UKQSAR meeting Sept 2023
Just got this message, the UKQSAR meetings are always excellent
We are pleased to announce that the next UKQSAR meeting will be held at the University of Liverpool on September 14th 2023.
The schedule will be: 09.00-10.00 Registration 10.00-16.00 Scientific programme
Please register using the online form: https://forms.office.com/e/RR5qGKGCwJ
We are delighted that the RSC CICAG group (Chemical Information and Computer Applications Group (rsccicag.org) have been able to offer two travel bursaries for students to attend the meeting in return for providing a short write-up for the group. If you would like to apply for the bursary please use the form: https://forms.office.com/e/yiNi4VFuZD
As usual, there is no cost to register for the meeting and we are grateful for MedChemica Ltd. for supporting the catering costs.
There will be plenty of opportunity for posters to be presented and discussed over lunch and coffee, please indicate if you would like to present a poster in the registration.
Information about travel options are available from the University of Liverpool website: https://www.liverpool.ac.uk/study/international/coming-to-liverpool/getting-here/ https://www.liverpool.ac.uk/maps/visiting/car-parking/ We would urge you to consider the environment when planning your journey.
ChEMBL 33 released
The latest update to ChEMBL has been released.
This fresh release comes with a few new data soures and also some new features: we added bioactivity data for understudied SLC targets from the RESOLUTE project and included a flag for Natural Products and for Chemical Probes. An annotation for the ACTIONTYPE of a measurement was included for approx. 270 K bioactivities. We also time-stamped every document in ChEMBL with their CREATIONDATE!
This version of the database, prepared on 31/05/2023 contains:
2,399,743 compounds (of which 2,372,674 have mol files)
3,051,613 compound records (non-unique compounds)
20,334,684 activities
1,610,596 assays
15,398 targets
88,630 documents
Full details are here http://chembl.blogspot.com/2023/06/release-of-chembl-33.html.
ChEMBL is a manually curated database of bioactive molecules with drug-like properties. It brings together chemical, bioactivity and genomic data to aid the translation of genomic information into effective new drugs.
Hot Topics: Targeting RNA 2023
A new initiative from the RSC BMCS. The BMCS Hot Topics online meetings are intended to highlight breaking areas of research in fields of science relevant to drug discovery. They will run as stand-alone half-day virtual events, 2-3 times per year.

The inaugural meeting on 30th November 2023 will cover relevant methods and modes of targeting RNA in drug discovery and will focus on how small molecules can accomplish this, including RNA binding and degradation, splicing modulation, and enzymes modifying RNA.
The first circular and registration can be found here https://www.rscbmcs.org/events/hottopicsrna23/.
Matthew Disney, University of Florida, Design of Bioactive Small Molecules Targeting RNA.
Elliott Bayle, Storm Therapeutics, Targeting RNA Modifying Enzymes: Successes, Challenges and Lessons Learned.
Maria Duca, Université Côte d’Azur – CNRS, Design of multifunctional conjugates for the targeting of non-coding RNAs: anticancer and antimicrobial applications.
Updated Hit identification page
The Hit Identification page to include information from a great analysis published recently. An Analysis of Successful Hit-to-Clinical Candidate Pairs?" DOI.

GARDP’s next webinar will take place on 7 June 2023
The latest Global Antibiotic Research & Development Partnership is on Project management in antimicrobial drug R&D.
Wed, Jun 7, 2023 12:30 PM - 2:00 PM BST
The webinar will feature the following speakers:
- Kristina Orrling, Programme Manager, Lygature, Netherlands
- Julie Miralves, R&D Portfolio and Planning Leader, Global Antibiotic R&D Partnership (GARDP), Switzerland
Register here https://register.gotowebinar.com/register/832555033649883487?source=network.
Detecting Bad Science with Data
A couple of interesting programs to listen to.
The More or Less team are highlighting an important area. https://www.bbc.co.uk/programmes/p0fpb87t.
For more than a decade there’ve been longstanding concerns about the credibility and reliability of science research. This “bad science” has often stemmed from poor data practice or worse. But statistics can also help us identify and understand some of what’s going wrong, whether that’s selective data-slicing or outright fabrication.
There is also a more detailed investigation on Radio 4 The Truth Police https://www.bbc.co.uk/programmes/m001lqvg.
For years, science has had a dirty secret; research has been dogged by claims and instances of fraud, malpractice and outright incompetence. Suspicious-looking data sets, breakthrough results that can’t be replicated, eyebrow-raising statistical sleights of hand, science has been undergoing something of an existential crisis.
This of course has a potential profound effect on drug discovery.
A recent study, reported in Science 28 August 2015: Vol. 349 no. 6251 DOI looking at psychological science, attempted to replicate published work suggests that 39% of effects replicated the original result. Also Amgen, tried to replicate 53 'landmark' cancer studies and failed to replicate the original studies in all but six occasions, Nature 483, 531–533 (29 March 2012) DOI.
A report by Arrowsmith noted that the success rates for new development projects in Phase II trials have fallen from 28% to 18% in recent years, with insufficient efficacy being the most frequent reason for failure (Phase II failures: 2008–2010. Nature Rev. Drug Discov. 10, 328–329 (2011))1. In a follow up article Nature Reviews Drug Discovery volume 10, page 712 (2011) it was reported that that only in ∼20–25% of the projects were the relevant published data completely in line with in-house findings. This resulted in extended target validation studies and in most cases project termination.
Target Validation
A couple of minor updates to the Target Validation page.
https://www.cambridgemedchemconsulting.com/resources/targetvalidation.html
Fake Publications in Biomedical Science
There have a number of headlines recently highlighting large language models (LLM https://en.wikipedia.org/wiki/Largelanguagemodel, most notably GTP-4 from OpenAI. These models are trained on vast amounts of data from a variety of sources and the quality of these data sources is not always as good as hoped.
It might be assumed the scientific literature would be of a higher standard but a recent preprint raises major concerns.
https://www.medrxiv.org/content/10.1101/2023.05.06.23289563v1
Fake Publications in Biomedical Science: Red-flagging Method Indicates Mass Production
Red-flagged fake publications (RFPs) account for around 28% of the published papers in biomedicine.
Wellcome Trust to triple size of Cambridge genome campus
Wellcome Britain’s biggest biomedical charity is to triple the size of its genetics and biological data facilities near Cambridge, in one of the largest investments in UK research infrastructure.
The site will continue to focus on genomics and biodata, aiming to attract global leaders in these fields and provide enhanced opportunities for this research to be translated into real-world solutions for health challenges. The new facilities will accommodate a range of occupiers from start-ups and scale-ups to more mature organisations, growing and enhancing the existing scientific ecosystem.
Anyone who has tried to travel down the A505 in the morning will also be interested in this item.
Upgrades to transport infrastructure, including to local road and cycle networks
Since the rail line goes right by the site could a station be included and better public transport access?
FDA Accepts Interim Analysis Plan for Ongoing Phase 2b Ibezapolstat Clinical Trial
FDA Accepts Interim Analysis Plan for Ongoing Phase 2b Ibezapolstat Clinical Trial and Acurx Announces Presentations at ECCMID 2023 Scientific Conference. This is an important step for Acurx, a great group of scientists to work with.
They will be presenting at the 33rd Annual European Congress of Clinical Microbiology and Infectious Disease (ECCMID) this month. Specifically, a scientific poster entitled "Novel pharmacology and susceptibility of ibezapolstat against C. difficile isolates with reduced susceptibility to C. difficile-directed antibiotics" will be presented by Dr. Kevin Garey, Professor and Chair, University of Houston College of Pharmacy and the Principal Investigator for microbiome aspects of our ibezapolstat clinical trial program.
Octopus trial for MS treatments
The Octopus trial is being led by researchers from the Queen Square MS Centre and the Medical Research Council (MRC) Clinical Trials Unit at University College London (UCL) and funded by the MS Society.
The multi-arm, multi-stage platform trial is designed to transform the way treatments for progressive MS are tested and will work up to three times faster than traditional trials.
More details…
https://www.ukri.org/news/participants-with-progressive-forms-of-ms-join-revolutionary-trial/.
More than 130,000 people live with MS in the UK and there are tens of thousands with progressive forms who have nothing to stop their MS getting worse. By tapping into the potential of approved drugs, which may have the potential to protect nerves, we can develop new treatments for MS faster.
Binding Sites are 3D
I've always found it interesting that whilst everyone recognises that protein binding sites are three dimensional (and chiral) there is a reluctance to have chiral centres in screening hits. This is despite examples were chiral centres aid affinity, selectivity and solubility. I suspect one of the concerns is the ease of follow up for any hits.
I've been working with Liverpool Chirochem to design a 3D rich, homochiral fragment screening library. The real beauty of this library is that the fragments can be easily expanded using validated chemistry in their parallel synthesis lab.
Once you have built a supply of these homochiral building blocks they can of course be put to many different additional uses, covalent fragments, DEL building blocks, and as building blocks for large virtual libraries. All of which can be supported by their parallel synthesis lab.
Cambridge MedChem Meeting
Registration for the SCI / RSC 22nd Medicinal Chemistry Symposium (better known as The Cambridge MedChem Meeting) 10 September - 13 September 2023 is now open.
As usual there are discounts available for RSC and SCI members, and RSC members need to use a discount code
RSC members should enter the Event discount code EJRFChem221 and select “Guest Member” under the section Member type. Delegates will be contacted for their RSC membership number after successful registration. RSC student members should enter the Event discount code EJRFChem221S and select “Guest Member” under the section Member type. Delegates will be contacted for their RSC membership number after successful registration.
You can register here https://www.soci.org/events/fine-chemicals-group/2023/sci--rsc-22nd-medicinal-chemistry-symposium.
As usual there is an outstanding lineup of international speakers.
As part of the conference, there is also the opportunity to participate in a Medicinal Chemistry Workshop which will take place on the Sunday afternoon.
ChEMBL 32 is released!
The fantastic resource ChEMBL has been updated. ChEMBL 32 contains
- 2,354,965 compounds (of which 2,327,928 have mol files)
- 2,995,433 compound records (non-unique compounds)
- 20,038,828 activities
- 1,536,903 assays
- 15,139 targets
- 86,364 documents
More details are here http://chembl.blogspot.com/2023/03/chembl-32-is-released.html.
Data can be downloaded from the ChEMBL FTP site.
Susceptibility testing in antibacterial drug R&D
This looks like an interesting webinar for all those interested in antibiotics research
Susceptibility testing in antibacterial drug R&D
Presentation 1: Pre-clinical antimicrobial susceptibility testing: considerations and challenges (Dee Shortridge):
- Steps for developing an in vitro susceptibility test for your lead compound
- Studies needed to characterize your compound
- Points to consider if your compound is not typical
Presentation 2: Pitfalls and opportunities of susceptibility testing in clinical trials of new antibiotics (Rafael Cantón):
- The introduction of new antimicrobials needs antimicrobial susceptibility testing to define their profile and the alignment with regulators.
- Susceptibility testing data are also used to define both clinical and PK/PD breakpoints.
- Moreover, they can be used to recognize wild type populations and anticipate emergence or resistance.
This is organised by the Global Antibiotic Research & Development Partnership (GARDP) https://gardp.org.
The Global Antibiotic Research & Development Partnership (GARDP) accelerates the development and access of treatments for drug-resistant infections. Together with private, public and non-profit partners, GARDP works to preserve the power of antibiotics for generations to come.
Updated Drug Discovery Resources
I have updated several pages in the Drug Discovery Resources.
Ester and Amide Bioisosteres https://www.cambridgemedchemconsulting.com/resources/bioisoteres/ester_bioisosteres.html
Cysteine Protease inhibitors https://www.cambridgemedchemconsulting.com/resources/hitidentification/focus/cysteineprotease_inhibitors.html Covalent Inhibitors https://www.cambridgemedchemconsulting.com/resources/lead_identification/covalent.html.
Annual site review
The Drug Discovery Resources website continues to be very popular with 161,922 page views. The pages were visited by over 76,200 viewers and around 20% of the visitors come back on multiple occasions suggesting they find it useful. The visitors come from 180 different countries with the top countries being
- United States (27%)
- United Kingdom (14%)
- India (10%)
- Germany (4%)
- South Korea (3%)
- China (3%)
One of the popular pages in 2021 was COVID-19 and the Identification of "Drug Candidates" a checklist for those using virtual screening to identify potential hits for COVID-19 targets. This has dropped out of the top pages in 2022.
The other most viewed pages were
- Lipophilicity
- Bioisosteres
- Molecular Interactions
- Distribution and Plasma Protein Binding
- Kinase Inhibitors
- Acid Bioisosteres
- ADME
- Aromatic Bioisosteres
- Aspartic Acid Proteases
- Solvation and desolvation
- HERG Activity
- Fragment based screening
Looking at the operating systems 54% are Windows users, 20% Mac users, 12% Android, 10% iOS and 2% Linux. The browsers of choice are Chrome and Safari with all other below 10%.
I don't know how comprehensive the analytics software is but there is approximately a 54:46 M:F split for Gender.