Seasons Greetings
It has been a mild winter here so far but some lovely still mornings. Take care everyone and have a great break and a successful New Year. As ever monies saved on cards will be donated to the MS Society. https://www.mssociety.org.uk.

Reproducibility Project: Cancer Biology
I've been waiting for this for a while. Reproducibility Project: Cancer Biology
The Reproducibility Project: Cancer Biology was an 8-year effort to replicate experiments from high-impact cancer biology papers published between 2010 and 2012. The project was a collaboration between the Center of Open Science and Science Exchange with all papers published as part of this project available in a collection at eLife and all replication data, code, and digital materials for the project available in a collection on OSF.
The work tried to repeat 193 experiments from 53 papers and found a significant number of challenges.

In summary
- Replication effect sizes were 85% smaller on average than the original findings
- 46% of effects replicated successfully on more criteria than they failed
- Original positive results were half as likely to replicate successfully (40%) than original null results (80%)
This quote from In the Pipeline is perhaps a useful reminder.
A robust result can probably be reproduced even if you switch to a different buffer, or if your cell lines have been passaged a different number of times, or if the concentration of the test molecule is a bit off, etc. The more persnickity and local the conditions have to be, the less robust your result is, and in general (sad to say) the lower the odds of it having a real-world impact in drug discovery. There are certainly important things that can only be demonstrated under very precise conditions, don’t get me wrong – but when you’re expecting umpteen thousand patients to take your drug candidate and show real effects, your underlying hypothesis needs to be able to take a good kicking and still come through.
EFMC awards
To acknowledge outstanding achievements in the field of Medicinal Chemistry and Chemical Biology, EFMC confers every two years three Awards. The 2022 Awards will be presented at the XXVII EFMC "International Symposium on Medicinal Chemistry" (EFMC-ISMC 2022) to be held in Nice, France on September 4-8, 2022.
All 3 awards consist of a diploma, 7.500€ and an invitation to give a headline presentation at EFMC-ISMC 2022.
THE NAUTA PHARMACOCHEMISTRY AWARD FOR MEDICINAL CHEMISTRY AND CHEMICAL BIOLOGY
THE UCB-EHRLICH AWARD FOR EXCELLENCE IN MEDICINAL CHEMISTRY
THE PROUS INSTITUTE - OVERTON and MEYER AWARD FOR NEW TECHNOLOGIES IN DRUG DISCOVERY
More details here https://www.efmc.info/awards
Nominations for these Awards consist of a nomination letter, a brief CV including a list of selected publications and two supporting letters. Self-nominations are also accepted. The nominations should be submitted to Prof. Rui Moreira, President of the EFMC here https://www.efmc.info/custawards.php?langue=english&clemenus=1187970296
Submission deadline: January 31, 2022
2022 Summer Placements at Diamond
Applications are now open for 12-week summer placements projects available at Diamond starting in June 2022. Absolutely fantastic opportunities.
https://www.diamond.ac.uk/Careers/Students/Summer-Placement/2022-Summer-Placements1.html
Diamond Light Source is the UK’s national synchrotron. It works like a giant microscope, harnessing the power of electrons to produce bright light that scientists can use to study anything from fossils to jet engines to viruses and vaccines.
Find out more about the European Lead Factory
On Tuesday 30 November, members of the European Lead Factory (ELF) will participate in a webinar organised by the Young Scientists Network (YSN) of the European Federation of Medicinal Chemistry and Chemical Biology (EFMC).
The topic of the session is “Biological Testing of Hit Compounds”. Dr Vera Nies, Programme Manager at Lygature (the coordinating partner of the ELF) will give an introduction to the European Lead Factory. Her talk will be followed by a presentation entitled "Best practices in High Throughput Screening: ELF as an example", by Dr Steven van Helden of Pivot Park Screening Centre. The last ELF member presentation will be given by Dr Phil Jones of BioAscent, who will talk about the “Approaches Towards PAINSless Lead Generation”.
More details and registration here https://www.europeanleadfactory.eu/newsroom/elf-participate-efmc-ysn-webinar
Medicines for millions of patients
A lot has happened over the last year but one of the high spots was the latest inductee to the 2020 RSC BMCS Hall of Fame. https://www.rscbmcs.org/awards/halloffame/
The BMCS is delighted to announce that David Rees PhD, FRSC, FMedSci, Chief Scientific Officer at Astex Pharmaceuticals, will be the 2020 inductee to its Hall of Fame, and the recipient of the associated medal. David is recognised internationally for his innovative use of chemistry in drug discovery. His research has forged bridges between academia and industry and he has held prominent positions in learned societies such as the Royal Society of Chemistry. He has led collaborations resulting in the discovery of three launched drugs, the anaesthetic agent Sugammadex which has been used in over 30 million patients in 60 countries, and the anti-cancer agents Ribociclib and Erdafitinib, both predicted to achieve blockbuster status. Much of Astex’s industry-leading productivity has been dependant on David’s chemical expertise. David is well known for his calm authority, scientific rigor and enthusiasm, and has over 140 publications and patents.
David gave a fabulous presentation at the 21st RSC / SCI Medicinal Chemistry Symposium in Cambridge and I'm delighted that this talk has been converted into an article published in RSC Medicinal Chemistry.
As many of you know I'm a keen advocate of looking at the number of patients treated rather than simple sales figures and David makes this point very eloquently.
ELF programs available for partnering
The European Lead Factory (ELF) is a collaborative public-private partnership aiming to deliver innovative drug discovery starting points. The ELFgives free access to up to 550,000 novel compounds, a unique industry-standard uHTS platform, and much more.
The success of the ELF approach has been widely acknowledged and the output has shown to be of high quality, worth following up and investment-ready. Funding for further development has been secured for several programmes, and by March 2018 two programmes have led to partnering deals being closed between the Programme Owner and an established pharmaceutical company.
A number of projects are now available for partnering.
The available projects are here.
Fluoxamine COVID-19 treatment
There have been several recent studies suggesting fluvoxamine may be beneficial when treating COVID-19 patients. Most of these reports are small trials that lack the statistical power to provide a solid answer. This has changed with the TOGETHER trial consortium who have been evaluating several different potential treatments shown in the table below. They also undertook a study investigating treatment with Ivermectin that was "stopped for futility".

The results of the Fluvoxamine trial have now been published "Effect of early treatment with fluvoxamine on risk of emergency care and hospitalisation among patients with COVID-19: the TOGETHER randomised, platform clinical trial" DOI. This appears to be a large (741 patients were allocated to fluvoxamine and 756 to placebo) well conducted blinded trial. The average age of the patients was 50 years, 42% male 58% female. Primary end points were related to hospitalisation. They enrolled only participants with diagnosed COVID-19 and less than 7 days of symptom onset using a commercially available COVID-19 rapid antigen test.
The proportion of patients observed in a COVID-19 emergency setting for more than 6 h or transferred to a teritary hospital due to COVID-19 was lower for the fluvoxamine group compared with placebo (79 [11%] of 741 vs 119 [16%] of 756).
The mechanism of action is uncertain but the initial hypothesis was based on the anti-inflammatory action of fluvoxamine. Fluvoxamine is a Selective serotonin reuptake inhibitor (SSRI), however if does have other activities CHEMBL1409/

SMILES = COCCCC/C(=N\OCCN)c1ccc(C(F)(F)F)cc1.O=C(O)/C=C\C(=O)O
It is worth noting Fluvoxamine is not an antiviral but it may act via anti-inflammatory effects that likely stem from its regulation of S1R, which modulates innate and adaptive immune responses and Fluvoxamine may serve to dampen cytokine storms. There have been reviews of the potential mechanism of action DOI.
Antiviral Molnupiravir Reduced the Risk of Hospitalization or Death by Approximately 50 Percent Compared to Placebo for Patients with Mild or Moderate COVID-19
Merck have just announced that the Investigational Oral Antiviral Molnupiravir Reduced the Risk of Hospitalization or Death by Approximately 50 Percent Compared to Placebo for Patients with Mild or Moderate COVID-19 in Positive Interim Analysis of Phase 3 Study.
Great news, but bear in mind this is only an interim study, so keep those fingers crossed.
Molnupiravir (development codes MK-4482 and EIDD-2801) is an experimental antiviral drug which is orally active and was developed for the treatment of influenza. It is a prodrug of the synthetic nucleoside derivative N4-hydroxycytidine, and exerts its antiviral action through introduction of copying errors during viral RNA replication.

InChI=1S/C13H19N3O7/c1-6(2)12(19)22-5-7-9(17)10(18)11(23-7)16-4-3-8(15-21)14-13(16)20/h3-4,6-7,9-11,17-18,21H,5H2,1-2H3,(H,14,15,20)/t7-,9-,10-,11-/m1/s1
CHEBI https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:180653
COVID moonshot to get £8M funding
A great start to the week.
The COVID Moonshot, a non-profit, open-science consortium of scientists from around the world dedicated to the discovery of globally affordable and easily-manufactured antiviral drugs against COVID-19 and future viral pandemics has received key funding of £8 million from Wellcome, on behalf of the Covid-19 Therapeutics Accelerator.
2022 RSC Medicinal Chemistry Emerging Investigator Lectureship
Nominations are now open for the 2022 RSC Medicinal Chemistry Emerging Investigator Lectureship and close on 08 October 2021. The lectureship is open to candidates who received their PhD in 2012 or later and who have made a significant contribution to medicinal chemistry in their early career.
The RSC welcome nominations from anyone, to be sent to our journal inbox at medchem-rsc@rsc.org, but ask that nominations include the name and affiliation of the researcher, along with at least one paragraph explaining their achievements and why they should be considered (a CV is not required, but is helpful if sent along with the nomination). Additionally, self-nominations are welcomed if accompanied by a letter of support from the nominees’ institute.
There are more details about nominations on our webpage here: https://www.rsc.org/journals-books-databases/about-journals/rsc-medicinal-chemistry/#Lectureship_MChemCom.
CYPlebrity: Machine learning models for the prediction of inhibitors of cytochrome P450 enzymes
CYPlebrity: Machine learning models for the prediction of inhibitors of cytochrome P450 enzymes DOI, https://nerdd.univie.ac.at/cyplebrity/, structures can be submitted in SMILES format or drawn using the sketcher. The calculation takes a few second per compound and the results are displayed as shown below.

Added to the section on CYP Interactions.
Alphafold in Opentargets
Thanks to fantastic work from the folks at UniProt, the Open Targets Platform target profile pages now feature DeepMind’s AlphaFold data.

Can be easily linked to disease associations.

AlphaFold Protein Structure Database
The AlphaFold Protein Structure Database Developed by DeepMind and EMBL-EBI is now available online.
AlphaFold DB provides open access to protein structure predictions for the human proteome and 20 other key organisms to accelerate scientific research.
AlphaFold DB currently provides predicted structures for the organisms listed below and includes human, laboratory species, and key pathogens. All the predictions for all the species can be downloaded from the EBI FTP site ftp://ftp.ebi.ac.uk/pub/databases/alphafold.
| Species | Common Name | Reference Proteome | Predicted Structures | Download |
|---|---|---|---|---|
| Arabidopsis thaliana | Arabidopsis | UP000006548 | 27,434 | Download (3642 MB) |
| Caenorhabditis elegans | Nematode worm | UP000001940 | 19,694 | Download (2601 MB) |
| Candida albicans | C. albicans | UP000000559 | 5,974 | Download (965 MB) |
| Danio rerio | Zebrafish | UP000000437 | 24,664 | Download (4141 MB) |
| Dictyostelium discoideum | Dictyostelium | UP000002195 | 12,622 | Download (2150 MB) |
| Drosophila melanogaster | Fruit fly | UP000000803 | 13,458 | Download (2174 MB) |
| Escherichia coli | E. coli | UP000000625 | 4,363 | Download (448 MB) |
| Glycine max | Soybean | UP000008827 | 55,799 | Download (7142 MB) |
| Homo sapiens | Human | UP000005640 | 23,391 | Download (4784 MB) |
| Leishmania infantum | L. infantum | UP000008153 | 7,924 | Download (1481 MB) |
| Methanocaldococcus jannaschii | M. jannaschii | UP000000805 | 1,773 | Download (171 MB) |
| Mus musculus | Mouse | UP000000589 | 21,615 | Download (3547 MB) |
| Mycobacterium tuberculosis | M. tuberculosis | UP000001584 | 3,988 | Download (421 MB) |
| Oryza sativa | Asian rice | UP000059680 | 43,649 | Download (4416 MB) |
| Plasmodium falciparum | P. falciparum | UP000001450 | 5,187 | Download (1132 MB) |
| Rattus norvegicus | Rat | UP000002494 | 21,272 | Download (3404 MB) |
| Saccharomyces cerevisiae | Budding yeast | UP000002311 | 6,040 | Download (960 MB) |
| Schizosaccharomyces pombe | Fission yeast | UP000002485 | 5,128 | Download (776 MB) |
| Staphylococcus aureus | S. aureus | UP000008816 | 2,888 | Download (268 MB) |
| Trypanosoma cruzi | T. cruzi | UP000002296 | 19,036 | Download (2905 MB) |
| Zea mays | Maize | UP000007305 | 39,299 | Download (5014 MB) |
The search bar at the top of the query page accepts queries based on protein name, gene name, UniProt identifier, or organism name. At present you can't search using a sequence and look for similar proteins. You would first need to do a BLAST search and use the results from that as queries.
Here I searched for Plasmodium falciparum carbonic anhydrase (Q8IHW5) a potential Malaria target. As you can see there is no crystal structure in the PDB. Whilst the active site is predicted with high confidence there are clearly regions for which there is very low confidence.

You can then download the structure in PDB or mmCIF format.
I made a homology model (in purple below) of this protein a while back and it has little sequence similarity with any proteins in the PDB. Despite not including a Zinc the Alphafold Predicted Structure includes histidines in positions to potentially coordinate to the Zinc. If it is possible to include the Zinc in the structure prediction I'd be interested in finding out.

Overall I'd say this is a very useful starting point.
The PROTACtable genome
As PROTACs have become more widespread the obvious question is which proteins are best suited to modulation by Protacs? A recent publication provides useful guidelines The PROTACtable genome DOI. The workflow is based on a method developed by a group at GSK, subsequently expanded and now integrated into the Open Targets Platform. Using publicly available data sources, the new method assesses whether a protein could be targeted using a PROTAC, based on the protein’s sequence, location, natural turnover rate in the cell, and evidence from published literature. The framework will help drug discovery researchers to gauge the PROTACtability of their protein of interest, and to prioritise their research accordingly.

More details on PROTACs here
AI4Proteins videos now online
On June 16/17 2021 RSC CICAG and AI3D held a joint meeting on Protein Structure Prediction. The full lineup of speakers, titles and abstracts can be found here.
Session 1: Session Chair: Professor Jeremy Frey (University of Southampton)
An AI solution to the protein folding problem: what is it, how did it happen, and some implications Professor John Moult (University of Maryland)
Session 2: Session Chair: Dr Melanie Vollmar (Diamond)
So you predicted a protein structure – What now? Dr Thomas Steinbrecher (Schrödinger)
Deep Learning enhanced prediction of protein structure and dynamics Dr Martina Audagnotto (AstraZeneca)
Fireflies-Lévy Flights algorithm for peptides conformational optimization Dr Zied Hosni (University of Sheffield)
Session 3: Session Chair: Dr Chris Swain (Cambridge MedChem Consulting)
How good are protein structure prediction methods at predicting folding pathways? Mr Carlos Outeiral Rubiera (University of Oxford)
Protein-Ligand Structure Prediction for GPCR Drug Design Dr Chris De Graaf (Sosei Heptares)
Session 4: Session Chair: Dr Márton Vass
Using icospherical input data in machine learning on the protein-binding problem Dr Ella Gale (University of Bristol)
Biological sequence design with machine learning Professor Debora Marks (Harvard University)
Session 5: Session Chair: Dr Simone Fulle (Novo Nordisk)
Lessons learned from generative models of biological sequences Professor Aleksej Zelezniak (Chalmers University of Technology)
DeepDock: a deep learning approach to predict ligand binding conformations Dr Oscar Méndez-Lucio (Janssen Pharmaceuticals)
Finding new in silico-based therapeutic strategies for IAHSP Dr Matteo Rossi Sebastiano (University of Turin)
Session 6: Session Chair: Professor Jonathan Goodman (University of Cambridge)
Designing molecular models by machine learning and experimental data Professor Cecilia Clementi (Freie Universität Berlin)
The “almost druggable” genome Professor Tudor Oprea (University of New Mexico)
Session 7: Session Chair: Dr Lucy Colwell (University of Cambridge)
General Effects of AI on Drug Discovery Dr Derek Lowe (Novartis)
Open Access Data: A Cornerstone for Artificial Intelligence Approaches to Protein Structure Prediction Professor Stephen Burley (RCSB PDB, Rutgers University, UCSD)
The videos of the presentations are now available on YouTube and you can access the playlist here https://www.youtube.com/playlist?list=PLBQwbn0mPhvWyTLnN6eFsbIwb5FByrs.
For those wanting a hype free insight into the impact AI might make on Drug Discovery then the presentation by Derek Lowe is well worth watching.
Open Targets Platform 21.06 has been released!
Open Targets Platform 21.06 has been released
The Open Targets Platform is a comprehensive tool that supports systematic identification and prioritisation of potential therapeutic drug targets. By integrating publicly available datasets including data generated by the Open Targets consortium, the Platform builds and scores target-disease associations to assist in drug target identification and prioritisation. It also integrates relevant annotation information about targets, diseases, phenotypes, and drugs, as well as their most relevant relationships.
Currently there are:-
Targets 60,606
Diseases 18,507
Drugs 13,185
Evidence strings 13,267,236
Associations 11,755,362
Computational Prediction of covalent Inhibitors
Covalent Inhibitors are an increasingly important class of therapeutic agents.
A computational pipeline has been described by the London lab to predict suggest covalent analogs of non-covalent ligands DOI.
Designing covalent inhibitors is increasingly important, although it remains challenging. Here, we present covalentizer, a computational pipeline for identifying irreversible inhibitors based on structures of targets with non-covalent binders. Through covalent docking of tailored focused libraries, we identify candidates that can bind covalently to a nearby cysteine while preserving the interactions of the original molecule. We found ∼11,000 cysteines proximal to a ligand across 8,386 complexes in the PDB. Of these, the protocol identified 1,553 structures with covalent predictions. In a prospective evaluation, five out of nine predicted covalent kinase inhibitors showed half-maximal inhibitory concentration (IC50) values between 155 nM and 4.5 μM. Application against an existing SARS-CoV Mpro reversible inhibitor led to an acrylamide inhibitor series with low micromolar IC50 values against SARS-CoV-2 Mpro. The docking was validated by 12 co-crystal structures. Together these examples hint at the vast number of covalent inhibitors accessible through our protocol.
RDKit was used for 2D molecular handling, conformation generation and RMSD calculation. RDKit: Open-source cheminformatics; version 2018.09.3; RDKit.org. Marvin was used in the process of preparing the molecules for docking, Marvin 17.21.0, ChemAxon (https://www.chemaxon.com). OpenBabel (http:// openbabel.org/wiki/Main_Page) was used to switch between molecular file formats. DOCKovalent (London et al., 2014) was used for virtual covalent docking. The Covalentizer code is available at https://github.com/LondonLab/Covalentizer.
Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2
An interesting open access paper in Science "Drug-induced phospholipidosis confounds drug repurposing for SARS-CoV-2" DOI points a potential flaw in interpreting in vitro data.
Repurposing drugs as treatments for COVID-19 has drawn much attention. Beginning with sigma receptor ligands, and expanding to other drugs from screening in the field, we became concerned that phospholipidosis was a shared mechanism underlying the antiviral activity of many repurposed drugs. For all of the 23 cationic amphiphilic drugs tested, including hydroxychloroquine, azithromycin, amiodarone, and four others already in clinical trials, phospholipidosis was monotonically correlated with antiviral efficacy. Conversely, drugs active against the same targets that did not induce phospholipidosis were not antiviral.
Phospholipidosis is well known phenomena for those involved in drug discovery, it is based on the physicochemical properties of the molecule and results in excessive accumulation of intracellular phospholipids in tissues, such as the liver, kidney and lung. The resulting accumulation can lead to liver, kidney, or respiratory failure.
Drug-induced phospholipidosis can be determined by measuring the accumulation of specific fluorescent probes in HepG2 cells or primary hepatocytes.
There is a more detailed explanation of the consequences here, "A Strategy for Risk Management of Drug-Induced Phospholipidosis" DOI.
6th RSC-BMCS Symposium on Mastering MedChem
The 6th RSC-BMCS symposium on mastering medicinal chemistry, a virtual event Tuesday & Wednesday, 29th & 30th June 2021 (two afternoon sessions).
This is a fantastic opportunity, two days of high quality presentations, the best way to learn MedChem is to listen to Case Histories. Details are here https://www.cathyhillevents.co.uk/BMCS/MMCVI.html.
You can register here https://www.maggichurchouseevents.co.uk/bmcs/MMCVI-online_registration.htm.

LifeArc funding Project Moonshot
The independent medical charity LifeArc is to support an international effort to rapidly develop a potential antiviral candidate for COVID-19.
The grant will be entirely dedicated to the COVID Moonshot initiative, which PostEra jointly leads alongside leading scientists from Diamond Light Source, Oxford University, Weizmann Institute, Memorial Sloan Kettering Cancer Center and MedChemica.
On the Antibacterial Action of Cultures of a Penicillium
May 10th 1929 is a very important day in drug discovery research, it was on this day that Alexander Fleming submitted his paper entitled "On the Antibacterial Action of Cultures of a Penicillium, with Special Reference to their Use in the Isolation of B. influenzæ" Br J Exp Pathol. 1929 Jun; 10(3): 226–236.

Over 90 years later this chance discovery still has a major impact on health today. Whilst isolation proved too challenging for Fleming he sent his Penicillium mold to anyone who requested it in hopes that they might isolate penicillin. It was only in 1940 that Howard Florey and team published the isolation and purification of Penicillin. Penicillin as a chemotherapeutic agent. Lancet. 1940;236:226–8. 10.1016/S0140-6736(01)08728-1.

The basic structure of the beta-lactam ring is shown below.

The beta lactam antibiotics (Penicillin and Cephalosporins) are a very well studied class of therapeutic agent, the mechanism of action is the inhibition of cell wall synthesis. Penicillin inhibits the formation of peptidoglycan cross-links in the bacterial cell wall; this is achieved through reaction of the β-lactam ring of penicillin to the enzyme DD-transpeptidase. As a consequence, DD-transpeptidase cannot catalyze formation of the cell wall cross-links.

Predicting sites of metabolism
I've updated the predicting sites of metabolism page
https://www.cambridgemedchemconsulting.com/resources/ADME/predicting_metabolism.html

Funding for novel antibiotic development
Registration is open for a GNA NOW #webinar taking place on Wednesday 12 May from 10:00-11:00 (CEST).
The GNA NOW Consortium is looking for a novel antibiotic compound to progress to a clinical candidate. To find out more about what the Consortium is looking for, what’s in it for you as a researcher, and how to submit a proposal, make sure not to miss this webinar! The session will feature presentations by Kristina Orrling from Lygature and Eric Bacqué of Evotec.
Register for free https://www.lygature.org/registration-gna-now-open-call-webinar
Funded by the @Innovative Medicines Initiative (IMI)
European Lead Factory call for proposals
The closing date for submissions to the European Lead Factory is Friday 28th May 2021.
Following the UK’s exit from the European Union on 31 January, we are pleased to confirm that the European Lead Factory still welcomes screening proposals from UK researchers.
This is a fantastic opportunity for researchers to get access to a top class high-throughput screening platform.

On this page, you can find the application forms, legal documents and all other relevant information required for your proposal submission.
The screening deck consists of around 535,000 compounds consisting of about 300,000 compounds from the original pharmaceutical companies, along with almost 190,000 compounds from custom design and synthesis. Furthermore, Grünenthal and Institut de Recherche Servier have provided around 50,000 new compounds not previously available to the European Lead Factory. Compounds from partners were selected for inclusion in the library based on several important quality criteria, as well as on the prerequisite that they are unique and not commercially available. Efforts were also taken to increase diversity, resulting in a top-quality compound library.
KLIFS kinase database
KLIFS is a kinase database that dissects experimental structures of catalytic kinase domains and the way kinase inhibitors interact with them. The KLIFS structural alignment enables the comparison of all structures and ligands to each other. Moreover, the KLIFS residue numbering scheme capturing the catalytic cleft with 85 residues enables the comparison of the interaction patterns of kinase-inhibitors, for example, to identify crucial interactions determining kinase-inhibitor selectivity. DOI.
Statistics
Kinases | 312 |
| Structures (# PDBs) | 5568 |
| Monomers | 12151 |
| Unique ligands | 3497 |
| KLIFS users in Apr-2021 | 897 |
Open Targets Platform: rebuilt, redesigned, reimagined
The Open Targets Platform has been updated.
- The new design of the Platform improves user experience and access to information. It has been reimagined to facilitate the building of new therapeutic hypotheses;
- New features include data on binary molecular interactions, and black box warnings;
- A complete refactoring of the codebase enables rapid development and new deployment strategies.
21st RSC / SCI Medicinal Chemistry Symposium
Registration for the 21st RSC / SCI Medicinal Chemistry Symposium Monday-Wednesday, 13th-15th September 2021 hosted at Churchill College, Cambridge, UK is now open. Twitter hashtag - #CamMedChem21
As usual there is a stella lineup of presentations and there is still time for some submissions.
I'd like to highlight one talk that is close to my heart. Ed Griffen will talking about The COVID Moonshot: SARS-CoV2 oral antiviral therapeutics from an Open Science global collaboration.
The COVID Moonshot is an ambitious crowdsourced initiative to accelerate the development of a COVID antiviral. We work in the open with no intellectual property constraints. This way, any scientist can view submitted drug designs and experimental data to inspire new design ideas. We use our cutting-edge machine learning tools and Folding@home's crowdsourced supercomputer to determine which drug designs to send to our partners to make and test in the lab. With each drug design tested, we get closer to our goal.
This talk will be in the late breaker session and because of the open nature of the project it will be a chance to really see the very latest results "hot off the press".
You can have a look at the latest results here now. https://covid.postera.ai/covid.
30th April: Late breaker and early poster deadline
23rd July: Final poster deadline
Registration link https://www.maggichurchouseevents.co.uk/bmcs/cmc21/index.htm
Do you include chiral fragments in your screen?
Fragment-based screening has become increasingly popular over the last 15 years and has proven to be a viable alternative to high-throughput screening.
A little while ago I posed the question "Do you include chiral fragments in your screen?" And the results were interesting. Whilst many, many folks clicked on the link very few actually responded to the poll, perhaps something people are thinking about but don't have an answer?
For those that did respond it seems that very few actively exclude chiral fragments.

Of those that included chiral fragments the responses were pretty evenly split between those that include racemates and only worry about the enantiomers if they appear as a hit, those that only include racemates if the individual enantiomers are available and those that only include individual enantiomers.
One of the arguments against including chiral fragments has been it increases the complexity of the fragments and as the ligand/receptor match becomes more complex the probability of any given molecule matching falls. However, biological targets are almost invariably chiral and selectivity often relies on stereochemical features.
One of the key attractions of fragment screening is the ability to explore/validate hits without committing significant chemistry resources, so I’d argue that the ready availability of related analogues ought to be part of the library selection criterion and by extension having access to the individual enantiomers should be an important consideration.
Chemical Probes
A really interesting review of the Chemical Probes portal
2020 was the first year of visible activity on the Chemical Probes Portal since 2017, with 115 probes added and over 500 compounds now included on the Portal. To celebrate, we’re highlighting ten of the best probes added to the Portal and evaluated by our Scientific Advisory Board in 2020. These probes are selective, potent, cell-active molecules that are rated four stars for use in cells and target new proteins or have new mechanisms of action. They include probes for previously ‘undruggable’ cancer targets, compounds that target GPCRs, epigenetic modulators and PROTACs.
Full details are here https://www.chemicalprobes.org/news/2020s-top-probes.

Open Targets Platform beta test
If you have a little time why not drop by the Open Targets website and give the beta test version and give them some feedback. Full details are in the blog post.
The beta version makes the most of the data from the recent 21.02 release and you can also interrogate the data using the brand new GraphQL API.
In particular, this version features:
- Redesigned evidence pages
- Updated drug profile pages
- More complete disease profile pages
Open Targets is a public-private partnership that uses human genetics and genomics data for systematic drug target identification and prioritisation. The current focus is on oncology, immunology and neurodegeneration.
Generating and interpreting the data required to identify a good drug target demands a diverse set of skills, backgrounds, evidence types and technologies, which do not exist today in any single entity. Open Targets brings together expertise from seven complementary institutions to systematically identify and prioritise targets from which safe and effective medicines can be developed.
March webinar: High Content Screening at the European Lead Factory
The next European Lead Factory webinar will take place on Thursday 11 March from 11:00-12:00 (CET). In this webinar, we will focus on High Content Screening with a presentation by Professor Jason Swedlow, head of the National Phenotypic Screening Centre at the University of Dundee.
More details and registration here https://www.europeanleadfactory.eu/node/375
MAIP: a web service for predicting blood‐stage malaria inhibitors
An interesting publication Bosc, N., Felix, E., Arcila, R. et al. MAIP: a web service for predicting blood‐stage malaria inhibitors. J Cheminform 13, 13 (2021). https://doi.org/10.1186/s13321-021-00487-2.
We describe the development of an open-source software platform for creating such models, a comprehensive evaluation of methods to create a single consensus model and a web platform called MAIP available at https://www.ebi.ac.uk/chembl/maip/. MAIP is freely available for the wider community to make large-scale predictions of potential malaria inhibiting compounds. This project also highlights some of the practical challenges in reproducing published computational methods and the opportunities that open-source software can offer to the community.
The code to standardise the compounds and train the models is available on GitHub:
standardiser: https://github.com/flatkinson/standardiser.
ModifiedNB: https://github.com/chembl/ModifiedNB.
mmv_train_image: https://github.com/chembl/mmv_train_image.
The ready to use Docker image to generate the models is available at https://hub.docker.com/r/chemblgroup/mmv_train. The MAIP platform is available at https://www.ebi.ac.uk/chembl/maip/.
New GARDP webinars

25 February: 'From discovery to IND: Roadmap to a successful antibacterial project' with Patricia Bradford and Alita Miller, moderated by Michael Mourez. Register here: https://attendee.gotowebinar.com/register/8974462807057685772?source=spark
4 March: 'Learning from COVID-19 to tackle the silent pandemic of antibiotic resistance' with Marc Mendelson, Joanne Liu and Manica Balasegaram. Register here: https://attendee.gotowebinar.com/register/3151890131955239691?source=spark
24 March: 'Discovering and developing new treatments for tuberculosis' with Nader Fotouhi, moderated by Lydia Nakiyingi. Register here: https://attendee.gotowebinar.com/register/394450209091477264?source=spark
As always, keep an eye on our website https://revive.gardp.org/webinars to find new webinar announcements and recordings of previous webinars.
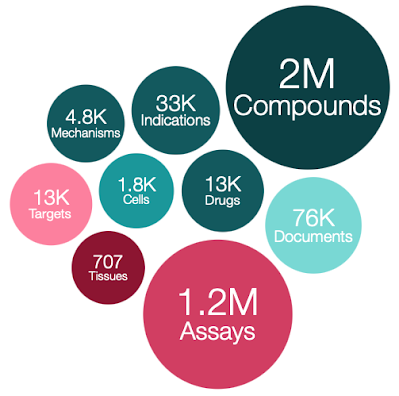
ChEMBL 28 has been released.
I see that a new version of ChEMBL has been released. Chembl 28
- 2,680,904 compound records
- 2,086,898 compounds (of which 2,066,376 have mol files)
- 17,276,334 activities
- 1,358,549 assays
- 14,347 targets
- 80,480 documents
Career choices when faced with changes of life
As someone who had to make a major career change due to personal circumstances I really recommend this sort of webinar that has been organised by the RSC.
Career choices when faced with changes of life
Join Career Consultants Cath Elmer and John Toscano who will spend an evening helping you give your working lives a well-deserved health check! Together we will look at some of the reasons career circumstances can change - out of choice or otherwise. We will discuss some of the career theories behind our decision-making processes to help us assess where we are and be better prepared for the future. This interactive event will be of interest whatever stage of your career.
Thursday 15th April 6:00-8:00 pm
Link https://www.rsc.org/events/detail/46366/career-choices-when-faced-with-changes-of-life.
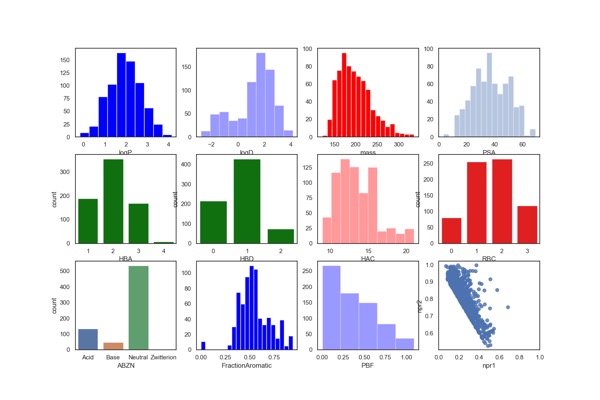
Fluorine Fragment Library
Key Organics have expanded the BIONET Fluorine Fragment Library which now includes 719 fluorinated fragments. All 719 fragments in the Fluorine Fragment Library have been analysed by 19F NMR and 1H NMR for:
- Structure verification
- Purity
- Measured solubility in PBS buffer ≥ 100μM
- Aggregation.
The calculated physicochemical properties of the library are shown below. They have also been filtered using PAINS and Lilly MedChem rules.
There are more Fragment Collections described here.
Predicting sites of metabolism
I've added GLORYx to the predicting metabolism page.
GLORYx predicts phase I and phase II metabolites for the chemical compound(s) provided by the user. The method is based on the FAME site of metabolism (SoM) prediction combined with sets of reaction rules encoding both phase I and phase II metabolic reactions.

I tried a range of other molecules and GLORYx was really very impressive in identifying potential metabolites.
Translational misconceptions
A really insightful and sobering article on the process of translating a basic research finding into a therapeutic product DOI, well worth a read and sharing.
By highlighting only the success stories, and using words like "pipeline" are we giving the false impression that there is a direct relationship between scientific discovery and novel drug, and the translation of one into the other is an inevitable and reliable process?
Chiral Fragment Screening
One of the attractions of fragment-based screening is that “Fragment Space” is smaller than “Chemical Space” and can be more effectively probed with a relatively small library. However, if we include chiral fragments the size of the library increases, as does the complexity of the ligands. It can also add extra work if the individual enantiomers of a chiral fragment are not available.
The poll linked below is intended to try and capture the current view. I'll post the results here later.
The Chemical Probes Portal now includes several PROTACs
The Chemical Probes Portal now includes several PROTACs in the list of recommended chemical probes and even more are under review by the Scientific Advisory Board.
PROTACs are bifunctional molecules that bind to the target protein and an E3 ligase, the simultaneous PROTAC binding of two proteins brings the target protein in close enough proximity for polyubiquitination by the E2 enzyme associated to the E3 ligase, which flags the target protein for degradation through the proteasome.

They have also put together a really useful list of criteria to help define the best choice of Protac probe.
David Rees BMCS Hall of Fame and Medal 2020
A lot has happened over the last couple of months but I wanted to highlight the latest inductee to the 2020 RSC BMCS Hall of Fame. https://www.rscbmcs.org/awards/halloffame/
The BMCS is delighted to announce that David Rees PhD, FRSC, FMedSci, Chief Scientific Officer at Astex Pharmaceuticals, will be the 2020 inductee to its Hall of Fame, and the recipient of the associated medal. David is recognised internationally for his innovative use of chemistry in drug discovery. His research has forged bridges between academia and industry and he has held prominent positions in learned societies such as the Royal Society of Chemistry. He has led collaborations resulting in the discovery of three launched drugs, the anaesthetic agent Sugammadex which has been used in over 30 million patients in 60 countries, and the anti-cancer agents Ribociclib and Erdafitinib, both predicted to achieve blockbuster status. Much of Astex’s industry-leading productivity has been dependant on David’s chemical expertise. David is well known for his calm authority, scientific rigor and enthusiasm, and has over 140 publications and patents.
The current pandemic means that the formal presentation has been delayed but I'm sure many will want to join me in congratulating David on this well-deserved prestigious award.
The single most important factor determining the likelihood of success of a project is the quality of the starting lead
Have you identified a disease mechanism that should be explored in a drug discovery project? Then you may have also considered developing a screening assay to find new chemical starting points that could serve as candidates for drug development.
In the next European Lead Factory webinar, taking place on Thursday 28 January, Dr Saman Honarnejad, Director of Drug Discovery at Pivot Park Screening Centre, will give a presentation on guidelines for HTS assay development, with an emphasis on methods to design, evaluate and improve conditions for biochemical and cellular assay formats.
The event will take place via Microsoft Teams on 28 January 2021 from 11:00-12:00 (CET). Discussions with the audience will form a key part of the webinar and time will be made available for questions and answers.
Registration is free using this link https://events.europeanleadfactory.eu/elf/elf-webinar/.
The European Lead Factory welcomes drug targets in all therapeutic areas. Interested? Submit your screening proposal now. The next deadline to apply will be in January 2021. Full details are on the ELF website.
I've been involved in a number of screening campaigns and I can confirm this is a high quality screening deck.
Annual Site Review
As 2021 starts I'd like to wish you all a Happy New Year and hope that 2021 marks the start of the recovery.
The Drug Discovery Resources website continues to increase in popularity with 147,000 page views, an increase of 5% over the figure for 2019. The pages were visited by over 76,000 viewers and around 20% of the visitors come back on multiple occasions suggesting they find it useful. The visitors come from 177 different countries with the top countries being
- United States (28%)
- United Kingdom (14%)
- India (10%)
- Germany (3%)
- Canada (3%)
- Japan (2.5%)
Perhaps not unexpectedly one of the popular pages was COVID-19 and the Identification of "Drug Candidates" a checklist for those using virtual screening to identify potential hits for COVID-19 targets.
The other most viewed pages were
- Lipophilicity
- Calculating Physicochemical Properties
- Bioisosteres
- Distribution and Plasma Protein Binding
- Molecular Interactions
- Kinase Inhibitors
- Acid Bioisosteres
- Aromatic Bioisosteres
- Aspartic Acid Proteases
- Solvation and desolvation
- HERG Activity
- Fragment based screening
- Protacs
Looking at the operating systems 54% are Windows users, 20% Mac users, 12% Android, 10% iOS and 2% Linux.

