CSD, ChEMBL, PDBe now interlinked
Three critical databases for drug discovery are now interlinked. The Cambridge Structural Database (CSD) a curated repository of small molecule crystal structures, ChEMBL a manually curated database of bioactive molecules with their associated biological data and PDBe a founding member of the Worldwide Protein Data Bank (wwPDB) which collects, organises and disseminates data on biological macromolecular structures, are now interlinked.
The BioChemGraph (BCG) project tackles the challenge of linking diverse data in biology by creating a resource that integrates data from the PDBe, ChEMBL, and the CSD. This has been achieved by mappings UniProt Accession ID and compound InChIKey, linkingmore than 17,000 experimentally determined protein-ligand complexes from the PDB to about 39,000 ChEMBL bioactivity records. By providing this link it is possible to not only identify binding affinity for the selected target but also much more information about the small molecule ligand, such as off-target activities, calculated physicochemical properties and also any ADME/T data that might be available. All data can be downloaded as a tsv as shown below.
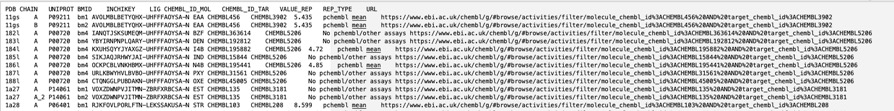
ChEMBL and PDBe have collaborated to set up an automatic pipeline for generating these data. As a result, the data will be updated weekly, in sync with the PDBe release every Wednesday at 00.00 UTC.
InChis have also been used to interconnect with Cambridge Structural Database using UniChem, 235,000 CSD identifiers have been linked corresponding entries in UniChem, a “universal translator” for chemistry using InChIs to connect chemical structures and their identifiers across various databases. UniChem enables researchers to seamlessly access information about a specific molecule across a wide variety of data sources. There are currently 41 data sources (https://www.ebi.ac.uk/unichem/sources).