A Nature Focus published in May gave a comprehensive update of the current efforts to combat Malaria, an illness caused by a parasite called Plasmodium, which is transmitted via the bites of infected mosquitoes. In the human body, the parasites multiply in the liver, and then infect red blood cells.

Five species of the plasmodium parasite can infect humans; the most serious forms of the disease are caused by Plasmodium falciparum. As of 2006 it accounted for 91% of all 247 million human malarial infections and 90% of the deaths.
Whilst vaccination would appear to be an obvious target, infected individuals never develop full immunity perhaps because the parasites live inside cells, where they are largely hidden from the immune response.
The life cycle of the malaria parasite has been studies extensively, and is described in the image below taken from the Center for Disease Control. The malaria parasite life cycle involves two hosts. During a blood meal, a malaria-infected female Anopheles mosquito inoculates sporozoites into the human host ![]() . Sporozoites infect liver cells
. Sporozoites infect liver cells ![]() and mature into schizonts
and mature into schizonts ![]() , which rupture and release merozoites
, which rupture and release merozoites ![]() . (Of note, in P. vivax and P. ovale a dormant stage [hypnozoites] can persist in the liver and cause relapses by invading the bloodstream weeks, or even years later.) After this initial replication in the liver (exo-erythrocytic schizogony
. (Of note, in P. vivax and P. ovale a dormant stage [hypnozoites] can persist in the liver and cause relapses by invading the bloodstream weeks, or even years later.) After this initial replication in the liver (exo-erythrocytic schizogony ![]() ), the parasites undergo asexual multiplication in the erythrocytes (erythrocytic schizogony
), the parasites undergo asexual multiplication in the erythrocytes (erythrocytic schizogony ![]() ). Merozoites infect red blood cells
). Merozoites infect red blood cells ![]() . The ring stage trophozoites mature into schizonts, which rupture releasing merozoites
. The ring stage trophozoites mature into schizonts, which rupture releasing merozoites ![]() . Some parasites differentiate into sexual erythrocytic stages (gametocytes)
. Some parasites differentiate into sexual erythrocytic stages (gametocytes) ![]() . Blood stage parasites are responsible for the clinical manifestations of the disease.
. Blood stage parasites are responsible for the clinical manifestations of the disease.
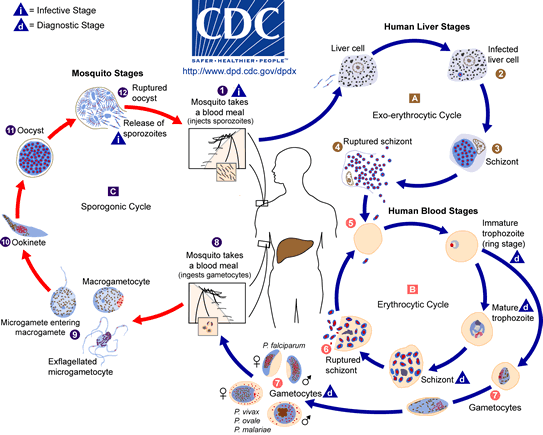
The gametocytes, male (microgametocytes) and female (macrogametocytes), are ingested by an Anopheles mosquito during a blood meal ![]() . The parasites’ multiplication in the mosquito is known as the sporogonic cycle
. The parasites’ multiplication in the mosquito is known as the sporogonic cycle ![]() . While in the mosquito's mid-gut, the microgametes penetrate the macrogametes generating zygotes
. While in the mosquito's mid-gut, the microgametes penetrate the macrogametes generating zygotes ![]() . The zygotes in turn become motile and elongated (ookinetes)
. The zygotes in turn become motile and elongated (ookinetes) ![]() which invade the midgut wall of the mosquito where they develop into oocysts
which invade the midgut wall of the mosquito where they develop into oocysts ![]() . The oocysts grow, rupture, and release sporozoites
. The oocysts grow, rupture, and release sporozoites ![]() , which make their way to the mosquito's salivary glands. Inoculation of the sporozoites into a new human host perpetuates the malaria life cycle
, which make their way to the mosquito's salivary glands. Inoculation of the sporozoites into a new human host perpetuates the malaria life cycle ![]() .
.
This understanding of the life-cycle will hopefully enable the identification of novel molecular targets.
High-throughput Screening
Recently two groups have reported the results of screening campaigns, GlaxoSmithKline (GSK) published ( doi:10.1038/nature09107) the results of screening nearly 2 million compounds from their chemical library for inhibitors of P. falciparum, of which 13,533 were confirmed to inhibit parasite growth by at least 80% at 2 uM. The chemical structures and associated data were made public to encourage additional drug lead identification efforts and further research into this disease. In a second paper (doi:10.1038/nature09099) a library containing 309,474 unique compounds, “designed at the scaffold level to provide diverse, comprehensive coverage of bioactive space”, was screened against Plasmodium falciparum strain 3D7 at a fixed concentration of 7 uM and afforded 1536 hits.
I thought I'd contribute to this generous effort by calculating a number of descriptors and properties for the molecules and then cluster the molecules in a variety of ways to help analysis and provide the data for download.
The supplementary data for both publications included all the structures in SMILES format, together with biological data and clustering using in house GSK tools. The Excel tables were exported as tab-delimitated text and then imported into a MOE database, the structures were subjected to the "wash" command to remove counterions and to check structures and protonation, the resulting structures were added to a new column in the MOE database and the "EXT_CMPD_NUMBER" added as the molecule name. This field was then exported in sdf (for_chemaxon.sdf) format for use with other tools. A variety of molecular properties were then calculated, and MACCS and PPP descriptors determined and the compounds clustered using the MACCS and PPP descriptors using a Tanimoto similarity index of 0.85. The clusters were then renumbered using this SVL script.
Several other programs were used to generate information, in particular the commandline tool cxcalc from ChemAxon was used to calculate pKa, LogD and LogP.
Commands used to generate ChemAxon data
Prompt$ cxcalc -i ID -o '/Users/swain/Desktop/Malaria/malaria_logd.txt' '/Users/swain/Desktop/Malaria/for_chemaxon.sdf' logd -H 7.4 logp
Prompt$ cxcalc -i ID -o '/Users/swain/Desktop/Malaria/malaria_pka.xt' '/Users/swain/Desktop/Malaria/for_chemaxon.sdf' pka -a 3 -b 3
A particularly useful method of clustering is to cluster by maximum common substructure (MCS), most chemists when looking a set of hits will endeavor to identify a common core structure and the automated clustering by MCS provides this sort of analysis. The MCS is the largest common part between two or more compounds excluding hydrogen atoms. A number of tools have been developed that offer clustering by MCS. I used LibMCS by ChemAxon they use similarity guided MCS, the idea being that compounds that are similar in structure are more likely to share MCS.
Prompt$ /Applications/ChemAxon/JChem/bin/libmcs /Users/swain/Desktop/Malaria/malaria.sdf -f -n 10 -r -o /Users/swain/Desktop/Malaria/malaria_mcs.sdf
These results were all then imported into the MOE database.
Physicochemical Profile of the results sets
A profile of the physicochemical properties was generated using this Applescript (RO5_script) that uses cxcalc to calculate the physicochemical properties and Aabel to constrict the histograms.
The set of histograms above show the profile of the compounds form the GSK (nature09099) screening effort, a significant number are relatively high molecular weight (>500) and high lipophilicity (clogP >5), and with so many high molecular weight compounds it is perhaps not surprising the rotatable bond count is relatively high. However there are still a large number of lower molecular weight compounds as potential leads.
The set of histograms below show the profile of the hits from the other screen (nature09107), as might be expected from a set of specifically designed libraries the logP and molecular weight show a much better profile with almost all MWt <500 and cLogP <5.
Comparison of Results sets
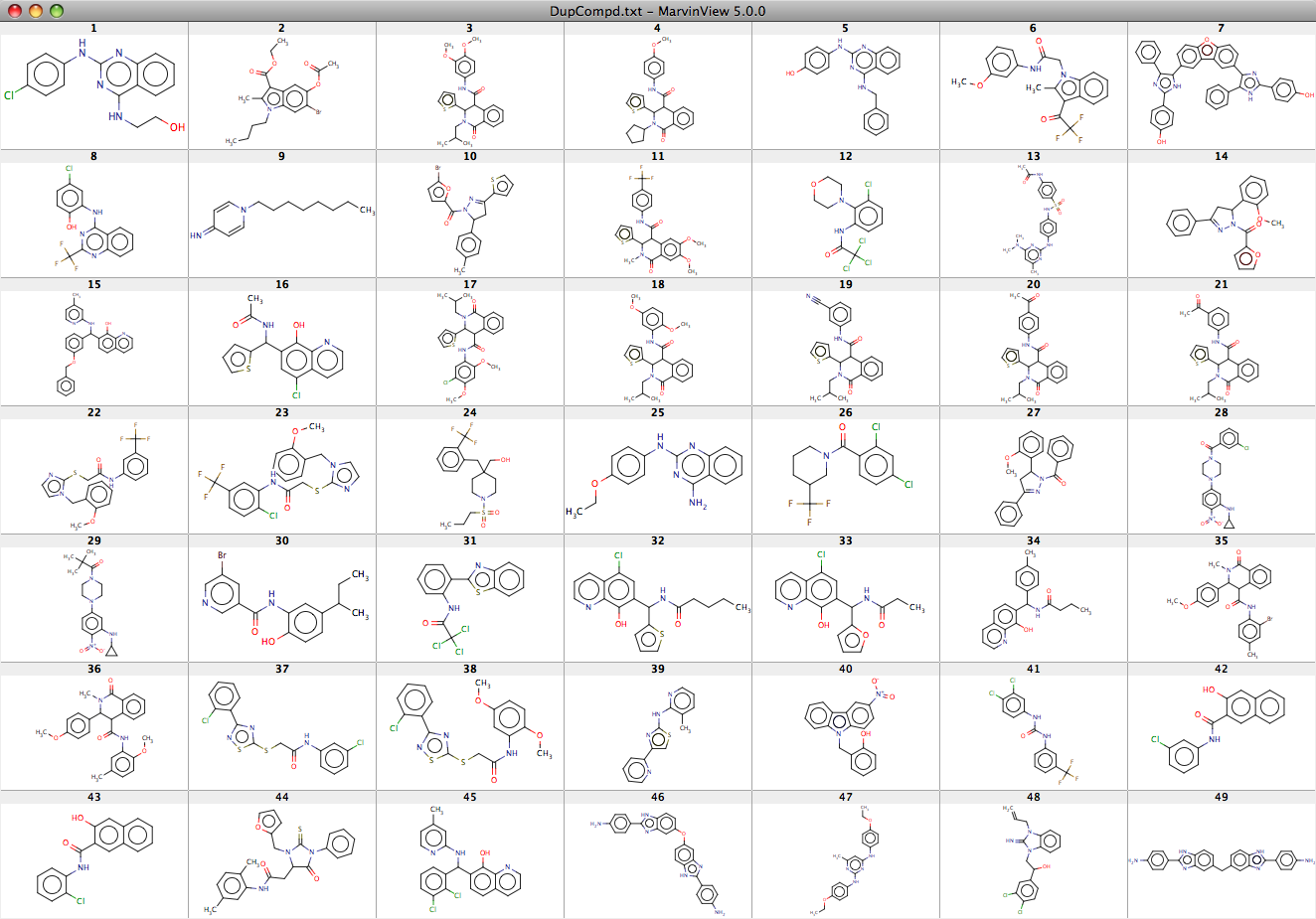
The two result sets were compared using a SVL script (db_nb_mols_incommon) that identifies identical compounds in multiple databases, there are only 49 identical compounds in the two results sets.
| Identical Compound Matrix | ||
| nature09107.mdb | nature09099.mdb | |
| nature09107.mdb | ||
| nature09099.mdb |
The 49 identical molecules are shown here as a screenshot taken from a MarvinView display.
Using the same script and running a similarity search identified compounds with a similarity (MaccsFP and Tan Coef 0.85), it is clear there is considerable overlap of similar compounds with approximately a third of the compounds from the nature09099 study having similar counterparts in the larger nature09107 study. This overlap gives confidence that these are not false positives, and can also offer some preliminary structure-activity information.
| Similar Compound Matrix | ||
| nature09107.mdb | nature09099.mdb | |
| nature09107.mdb | ||
| nature09099.mdb |
Next steps
First Page
Cluster Analysis
Using the Filemaker Pro Database
